Chufan Gao
Utilizing Training Data to Improve LLM Reasoning for Tabular Understanding
Aug 26, 2025Abstract:Automated tabular understanding and reasoning are essential tasks for data scientists. Recently, Large language models (LLMs) have become increasingly prevalent in tabular reasoning tasks. Previous work focuses on (1) finetuning LLMs using labeled data or (2) Training-free prompting LLM agents using chain-of-thought (CoT). Finetuning offers dataset-specific learning at the cost of generalizability. Training-free prompting is highly generalizable but does not take full advantage of training data. In this paper, we propose a novel prompting-based reasoning approach, Learn then Retrieve: LRTab, which integrates the benefits of both by retrieving relevant information learned from training data. We first use prompting to obtain CoT responses over the training data. For incorrect CoTs, we prompt the LLM to predict Prompt Conditions to avoid the error, learning insights from the data. We validate the effectiveness of Prompt Conditions using validation data. Finally, at inference time, we retrieve the most relevant Prompt Conditions for additional context for table understanding. We provide comprehensive experiments on WikiTQ and Tabfact, showing that LRTab is interpretable, cost-efficient, and can outperform previous baselines in tabular reasoning.
TrialSynth: Generation of Synthetic Sequential Clinical Trial Data
Sep 11, 2024


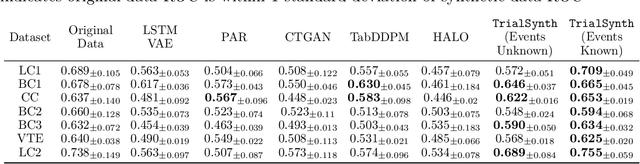
Abstract:Analyzing data from past clinical trials is part of the ongoing effort to optimize the design, implementation, and execution of new clinical trials and more efficiently bring life-saving interventions to market. While there have been recent advances in the generation of static context synthetic clinical trial data, due to both limited patient availability and constraints imposed by patient privacy needs, the generation of fine-grained synthetic time-sequential clinical trial data has been challenging. Given that patient trajectories over an entire clinical trial are of high importance for optimizing trial design and efforts to prevent harmful adverse events, there is a significant need for the generation of high-fidelity time-sequence clinical trial data. Here we introduce TrialSynth, a Variational Autoencoder (VAE) designed to address the specific challenges of generating synthetic time-sequence clinical trial data. Distinct from related clinical data VAE methods, the core of our method leverages Hawkes Processes (HP), which are particularly well-suited for modeling event-type and time gap prediction needed to capture the structure of sequential clinical trial data. Our experiments demonstrate that TrialSynth surpasses the performance of other comparable methods that can generate sequential clinical trial data, in terms of both fidelity and in enabling the generation of highly accurate event sequences across multiple real-world sequential event datasets with small patient source populations when using minimal external information. Notably, our empirical findings highlight that TrialSynth not only outperforms existing clinical sequence-generating methods but also produces data with superior utility while empirically preserving patient privacy.
Automatically Labeling $200B Life-Saving Datasets: A Large Clinical Trial Outcome Benchmark
Jun 13, 2024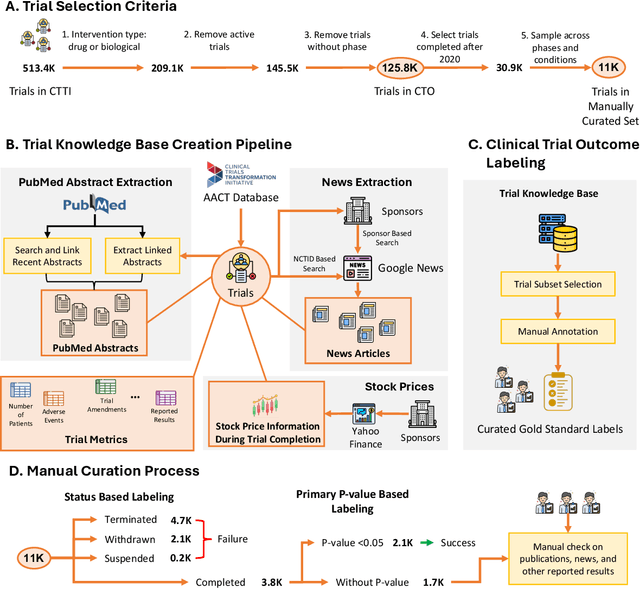
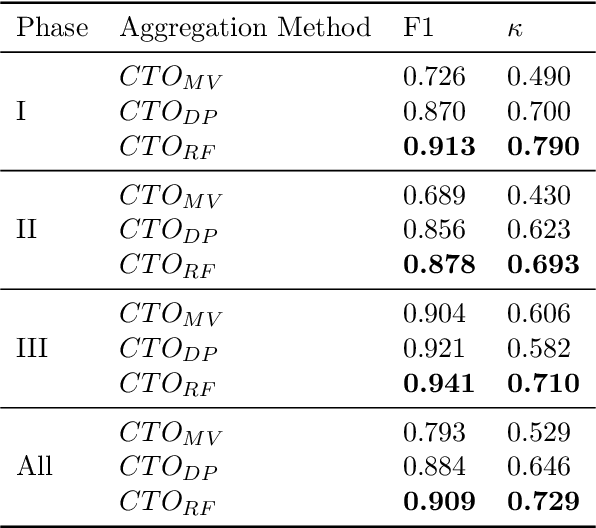
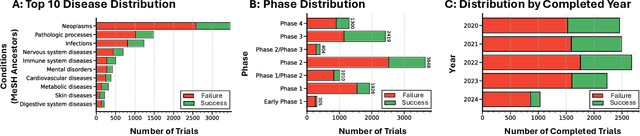
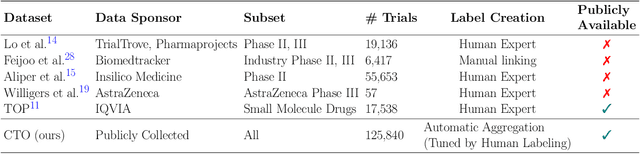
Abstract:The global cost of drug discovery and development exceeds $200 billion annually. The main results of drug discovery and development are the outcomes of clinical trials, which directly influence the regulatory approval of new drug candidates and ultimately affect patient outcomes. Despite their significance, large-scale, high-quality clinical trial outcome data are not readily available to the public. Suppose a large clinical trial outcome dataset is provided; machine learning researchers can potentially develop accurate prediction models using past trials and outcome labels, which could help prioritize and optimize therapeutic programs, ultimately benefiting patients. This paper introduces Clinical Trial Outcome (CTO) dataset, the largest trial outcome dataset with around 479K clinical trials, aggregating outcomes from multiple sources of weakly supervised labels, minimizing the noise from individual sources, and eliminating the need for human annotation. These sources include large language model (LLM) decisions on trial-related documents, news headline sentiments, stock prices of trial sponsors, trial linkages across phases, and other signals such as patient dropout rates and adverse events. CTO's labels show unprecedented agreement with supervised clinical trial outcome labels from test split of the supervised TOP dataset, with a 91 F1.
TTM-RE: Memory-Augmented Document-Level Relation Extraction
Jun 09, 2024Abstract:Document-level relation extraction aims to categorize the association between any two entities within a document. We find that previous methods for document-level relation extraction are ineffective in exploiting the full potential of large amounts of training data with varied noise levels. For example, in the ReDocRED benchmark dataset, state-of-the-art methods trained on the large-scale, lower-quality, distantly supervised training data generally do not perform better than those trained solely on the smaller, high-quality, human-annotated training data. To unlock the full potential of large-scale noisy training data for document-level relation extraction, we propose TTM-RE, a novel approach that integrates a trainable memory module, known as the Token Turing Machine, with a noisy-robust loss function that accounts for the positive-unlabeled setting. Extensive experiments on ReDocRED, a benchmark dataset for document-level relation extraction, reveal that TTM-RE achieves state-of-the-art performance (with an absolute F1 score improvement of over 3%). Ablation studies further illustrate the superiority of TTM-RE in other domains (the ChemDisGene dataset in the biomedical domain) and under highly unlabeled settings.
Language Interaction Network for Clinical Trial Approval Estimation
Apr 26, 2024



Abstract:Clinical trial outcome prediction seeks to estimate the likelihood that a clinical trial will successfully reach its intended endpoint. This process predominantly involves the development of machine learning models that utilize a variety of data sources such as descriptions of the clinical trials, characteristics of the drug molecules, and specific disease conditions being targeted. Accurate predictions of trial outcomes are crucial for optimizing trial planning and prioritizing investments in a drug portfolio. While previous research has largely concentrated on small-molecule drugs, there is a growing need to focus on biologics-a rapidly expanding category of therapeutic agents that often lack the well-defined molecular properties associated with traditional drugs. Additionally, applying conventional methods like graph neural networks to biologics data proves challenging due to their complex nature. To address these challenges, we introduce the Language Interaction Network (LINT), a novel approach that predicts trial outcomes using only the free-text descriptions of the trials. We have rigorously tested the effectiveness of LINT across three phases of clinical trials, where it achieved ROC-AUC scores of 0.770, 0.740, and 0.748 for phases I, II, and III, respectively, specifically concerning trials involving biologic interventions.
Towards Adapting Open-Source Large Language Models for Expert-Level Clinical Note Generation
Apr 25, 2024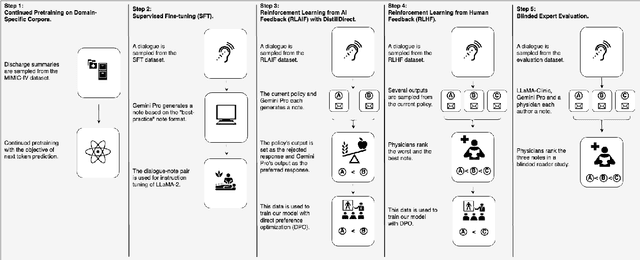
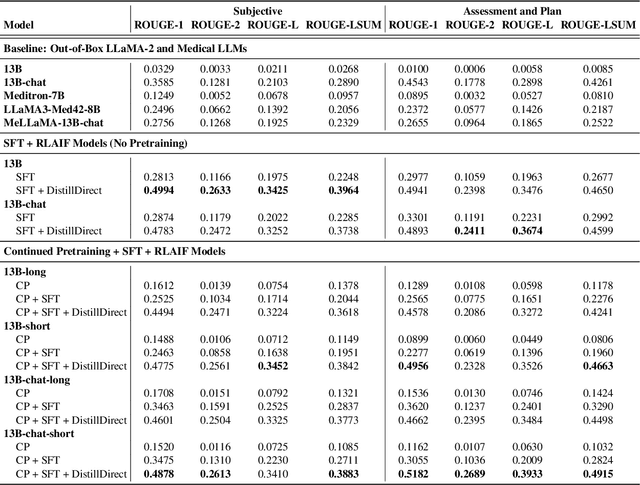
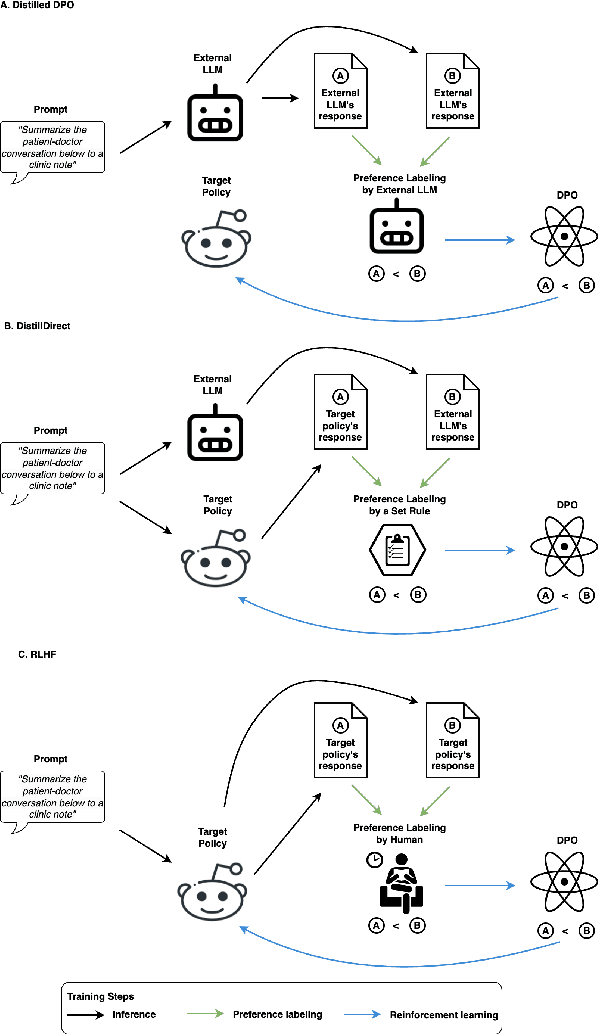
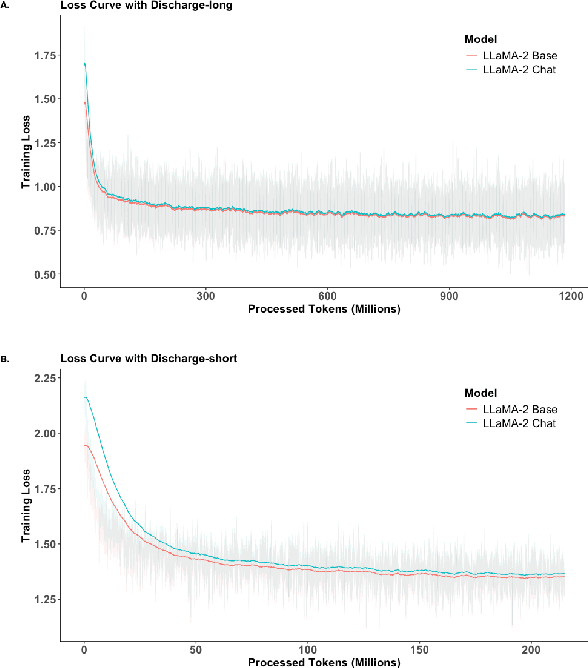
Abstract:Large Language Models (LLMs) have shown promising capabilities in handling clinical text summarization tasks. In this study, we demonstrate that a small open-source LLM can be effectively trained to generate high-quality clinical notes from outpatient patient-doctor dialogues. We achieve this through a comprehensive domain- and task-specific adaptation process for the LLaMA-2 13 billion parameter model. This process incorporates continued pre-training, supervised fine-tuning, and reinforcement learning from both AI and human feedback. We introduced an enhanced approach, termed DistillDirect, for performing on-policy reinforcement learning with Gemini Pro serving as the teacher model. Our resulting model, LLaMA-Clinic, is capable of generating clinical notes that are comparable in quality to those authored by physicians. In a blinded physician reader study, the majority (90.4%) of individual evaluations rated the notes generated by LLaMA-Clinic as "acceptable" or higher across all three criteria: real-world readiness, completeness, and accuracy. Notably, in the more challenging "Assessment and Plan" section, LLaMA-Clinic scored higher (4.2/5) in real-world readiness compared to physician-authored notes (4.1/5). Additionally, we identified caveats in public clinical note datasets, such as ACI-BENCH. We highlight key considerations for future clinical note-generation tasks, emphasizing the importance of pre-defining a best-practice note format. Overall, our research demonstrates the potential and feasibility of training smaller, open-source LLMs to assist with clinical documentation, capitalizing on healthcare institutions' access to patient records and domain expertise. We have made our newly created synthetic clinic dialogue-note dataset and the physician feedback dataset publicly available to foster future research in this field.
Signal Quality Auditing for Time-series Data
Feb 01, 2024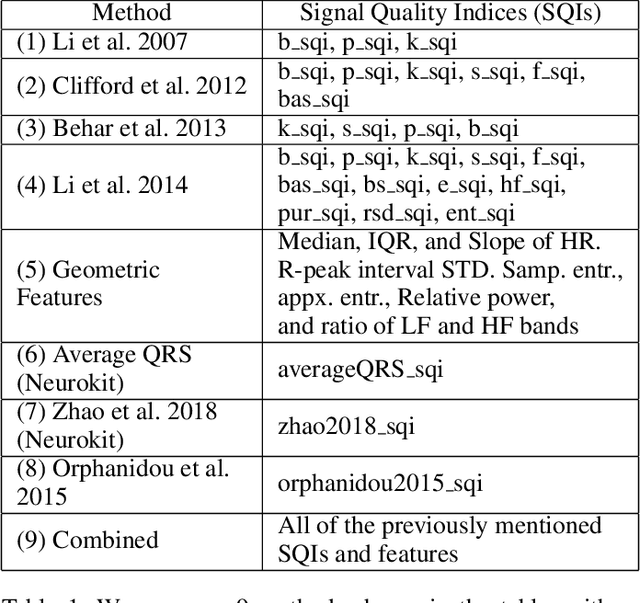
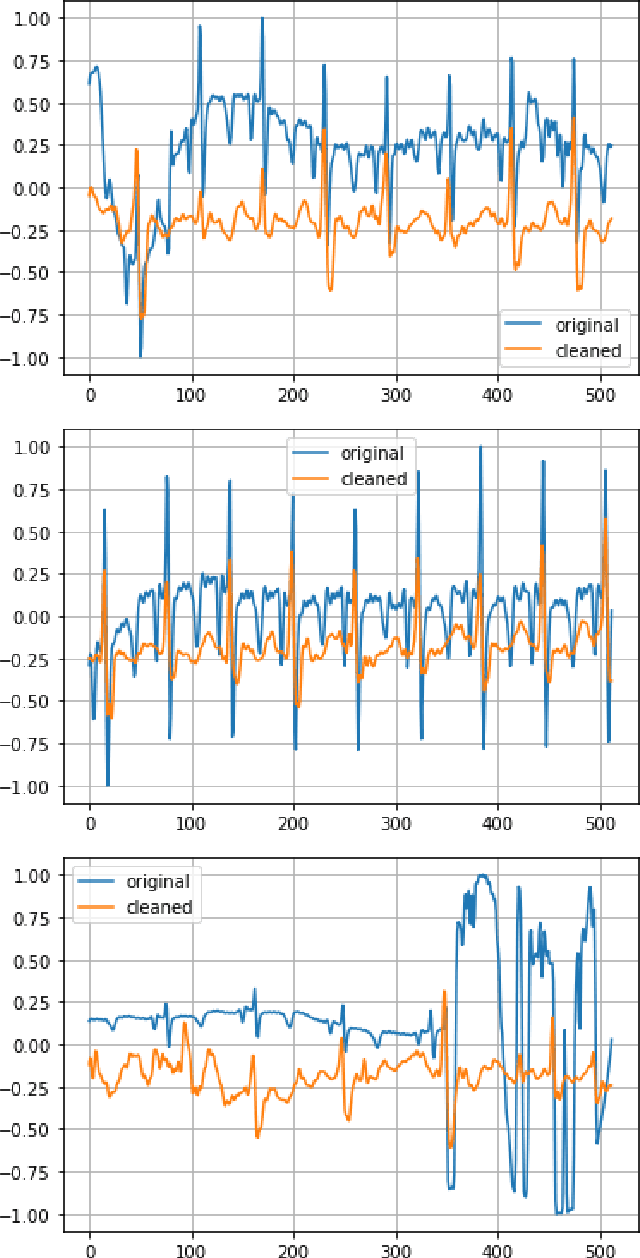
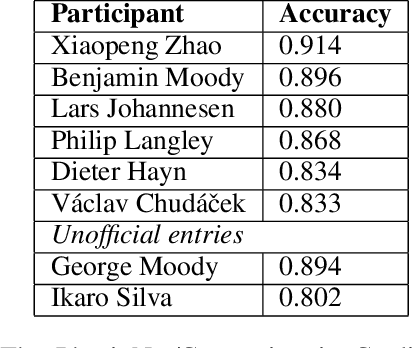
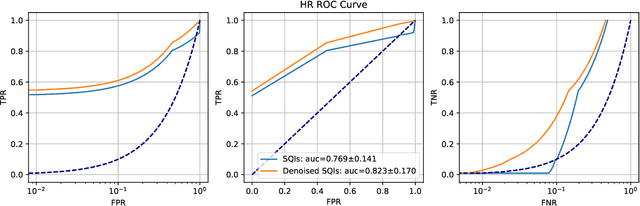
Abstract:Signal quality assessment (SQA) is required for monitoring the reliability of data acquisition systems, especially in AI-driven Predictive Maintenance (PMx) application contexts. SQA is vital for addressing "silent failures" of data acquisition hardware and software, which when unnoticed, misinform the users of data, creating the risk for incorrect decisions with unintended or even catastrophic consequences. We have developed an open-source software implementation of signal quality indices (SQIs) for the analysis of time-series data. We codify a range of SQIs, demonstrate them using established benchmark data, and show that they can be effective for signal quality assessment. We also study alternative approaches to denoising time-series data in an attempt to improve the quality of the already degraded signal, and evaluate them empirically on relevant real-world data. To our knowledge, our software toolkit is the first to provide an open source implementation of a broad range of signal quality assessment and improvement techniques validated on publicly available benchmark data for ease of reproducibility. The generality of our framework can be easily extended to assessing reliability of arbitrary time-series measurements in complex systems, especially when morphological patterns of the waveform shapes and signal periodicity are of key interest in downstream analyses.
PromptRE: Weakly-Supervised Document-Level Relation Extraction via Prompting-Based Data Programming
Oct 13, 2023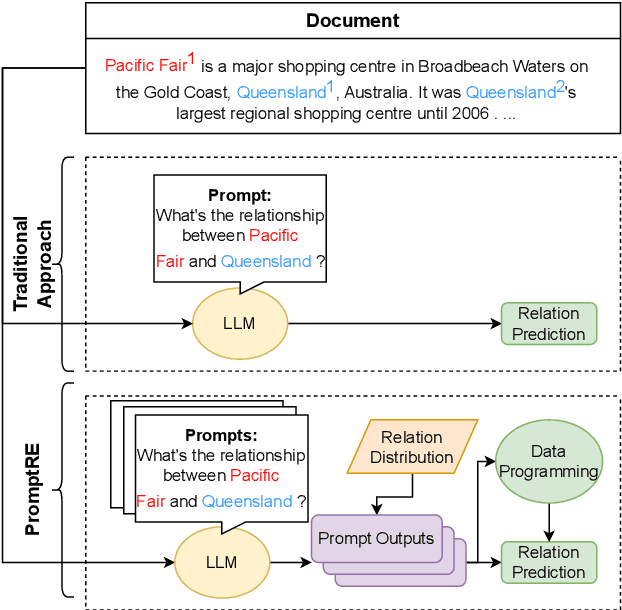



Abstract:Relation extraction aims to classify the relationships between two entities into pre-defined categories. While previous research has mainly focused on sentence-level relation extraction, recent studies have expanded the scope to document-level relation extraction. Traditional relation extraction methods heavily rely on human-annotated training data, which is time-consuming and labor-intensive. To mitigate the need for manual annotation, recent weakly-supervised approaches have been developed for sentence-level relation extraction while limited work has been done on document-level relation extraction. Weakly-supervised document-level relation extraction faces significant challenges due to an imbalanced number "no relation" instances and the failure of directly probing pretrained large language models for document relation extraction. To address these challenges, we propose PromptRE, a novel weakly-supervised document-level relation extraction method that combines prompting-based techniques with data programming. Furthermore, PromptRE incorporates the label distribution and entity types as prior knowledge to improve the performance. By leveraging the strengths of both prompting and data programming, PromptRE achieves improved performance in relation classification and effectively handles the "no relation" problem. Experimental results on ReDocRED, a benchmark dataset for document-level relation extraction, demonstrate the superiority of PromptRE over baseline approaches.
DRG-LLaMA : Tuning LLaMA Model to Predict Diagnosis-related Group for Hospitalized Patients
Sep 29, 2023



Abstract:In the U.S. inpatient payment system, the Diagnosis-Related Group (DRG) is pivotal, but its assignment process is inefficient. The study introduces DRG-LLaMA, an advanced large language model (LLM) fine-tuned on clinical notes to enhance DRGs assignment. Utilizing LLaMA as the foundational model and optimizing it through Low-Rank Adaptation (LoRA) on 236,192 MIMIC-IV discharge summaries, our DRG-LLaMA-7B model exhibited a noteworthy macro-averaged F1 score of 0.327, a top-1 prediction accuracy of 52.0%, and a macro-averaged Area Under the Curve (AUC) of 0.986, with a maximum input token length of 512. This model surpassed the performance of prior leading models in DRG prediction, showing a relative improvement of 40.3% and 35.7% in macro-averaged F1 score compared to ClinicalBERT and CAML, respectively. Applied to base DRG and complication or comorbidity (CC)/major complication or comorbidity (MCC) prediction, DRG-LLaMA achieved a top-1 prediction accuracy of 67.8% and 67.5%, respectively. Additionally, our findings indicate that DRG-LLaMA's performance correlates with increased model parameters and input context lengths.
AnyPredict: Foundation Model for Tabular Prediction
May 20, 2023Abstract:Foundation models are pre-trained on massive data to perform well across many downstream tasks. They have demonstrated significant success in natural language processing and computer vision. Nonetheless, the use of such models in tabular prediction tasks has been limited, with the main hurdles consisting of (1) the lack of large-scale and diverse tabular datasets with standardized labels and (2) the schema mismatch and predictive target heterogeneity across domains. This paper proposes a method for building training data at scale for tabular prediction foundation models (AnyPredict) using both in-domain and a wide range of out-domain datasets. The method uses a data engine that leverages large language models (LLMs) to consolidate tabular samples to overcome the barrier across tables with varying schema and align out-domain data with the target task using a ``learn, annotate, and audit'' pipeline. The expanded training data enables the pre-trained AnyPredict to support every tabular dataset in the domain without fine-tuning, resulting in significant improvements over supervised baselines: it reaches an average ranking of 1.57 and 1.00 on 7 patient outcome prediction datasets and 3 trial outcome prediction datasets, respectively. In addition, AnyPredict exhibits impressive zero-shot performances: it outperforms supervised XGBoost models by 8.9% and 17.2% on average in two prediction tasks, respectively.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge