Rakesh Mullick
Label Sharing Incremental Learning Framework for Independent Multi-Label Segmentation Tasks
Nov 17, 2024



Abstract:In a setting where segmentation models have to be built for multiple datasets, each with its own corresponding label set, a straightforward way is to learn one model for every dataset and its labels. Alternatively, multi-task architectures with shared encoders and multiple segmentation heads or shared weights with compound labels can also be made use of. This work proposes a novel label sharing framework where a shared common label space is constructed and each of the individual label sets are systematically mapped to the common labels. This transforms multiple datasets with disparate label sets into a single large dataset with shared labels, and therefore all the segmentation tasks can be addressed by learning a single model. This eliminates the need for task specific adaptations in network architectures and also results in parameter and data efficient models. Furthermore, label sharing framework is naturally amenable for incremental learning where segmentations for new datasets can be easily learnt. We experimentally validate our method on various medical image segmentation datasets, each involving multi-label segmentation. Furthermore, we demonstrate the efficacy of the proposed method in terms of performance and incremental learning ability vis-a-vis alternative methods.
One-shot Localization and Segmentation of Medical Images with Foundation Models
Oct 28, 2023



Abstract:Recent advances in Vision Transformers (ViT) and Stable Diffusion (SD) models with their ability to capture rich semantic features of the image have been used for image correspondence tasks on natural images. In this paper, we examine the ability of a variety of pre-trained ViT (DINO, DINOv2, SAM, CLIP) and SD models, trained exclusively on natural images, for solving the correspondence problems on medical images. While many works have made a case for in-domain training, we show that the models trained on natural images can offer good performance on medical images across different modalities (CT,MR,Ultrasound) sourced from various manufacturers, over multiple anatomical regions (brain, thorax, abdomen, extremities), and on wide variety of tasks. Further, we leverage the correspondence with respect to a template image to prompt a Segment Anything (SAM) model to arrive at single shot segmentation, achieving dice range of 62%-90% across tasks, using just one image as reference. We also show that our single-shot method outperforms the recently proposed few-shot segmentation method - UniverSeg (Dice range 47%-80%) on most of the semantic segmentation tasks(six out of seven) across medical imaging modalities.
Pristine annotations-based multi-modal trained artificial intelligence solution to triage chest X-ray for COVID-19
Nov 10, 2020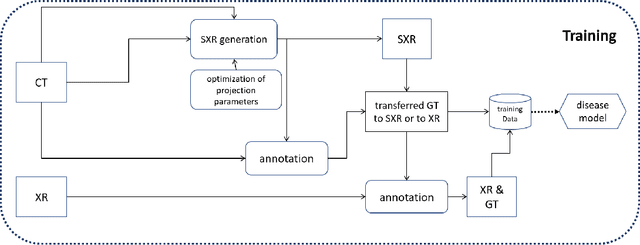



Abstract:The COVID-19 pandemic continues to spread and impact the well-being of the global population. The front-line modalities including computed tomography (CT) and X-ray play an important role for triaging COVID patients. Considering the limited access of resources (both hardware and trained personnel) and decontamination considerations, CT may not be ideal for triaging suspected subjects. Artificial intelligence (AI) assisted X-ray based applications for triaging and monitoring require experienced radiologists to identify COVID patients in a timely manner and to further delineate the disease region boundary are seen as a promising solution. Our proposed solution differs from existing solutions by industry and academic communities, and demonstrates a functional AI model to triage by inferencing using a single x-ray image, while the deep-learning model is trained using both X-ray and CT data. We report on how such a multi-modal training improves the solution compared to X-ray only training. The multi-modal solution increases the AUC (area under the receiver operating characteristic curve) from 0.89 to 0.93 and also positively impacts the Dice coefficient (0.59 to 0.62) for localizing the pathology. To the best our knowledge, it is the first X-ray solution by leveraging multi-modal information for the development.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge