Douglas N. Greve
Martinos Center for Biomedical Imaging, Boston, Harvard Medical School, Boston
Recon-all-clinical: Cortical surface reconstruction and analysis of heterogeneous clinical brain MRI
Sep 05, 2024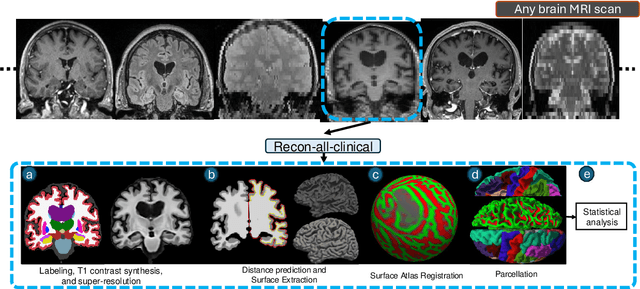
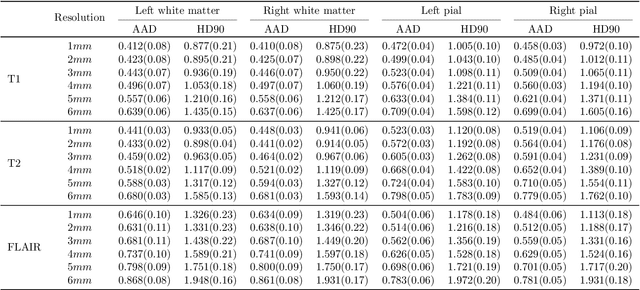
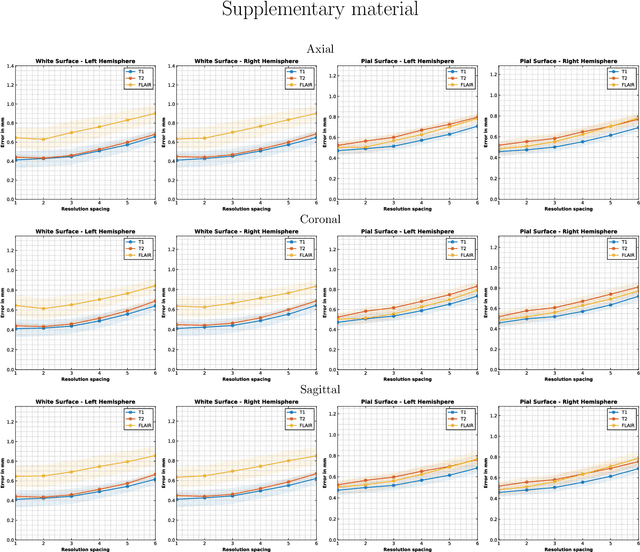
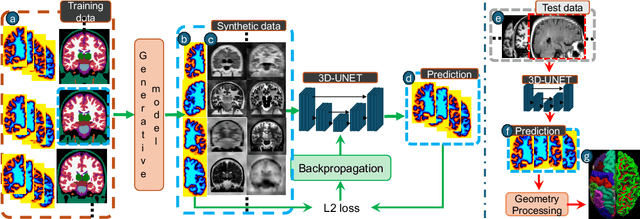
Abstract:Surface-based analysis of the cerebral cortex is ubiquitous in human neuroimaging with MRI. It is crucial for cortical registration, parcellation, and thickness estimation. Traditionally, these analyses require high-resolution, isotropic scans with good gray-white matter contrast, typically a 1mm T1-weighted scan. This excludes most clinical MRI scans, which are often anisotropic and lack the necessary T1 contrast. To enable large-scale neuroimaging studies using vast clinical data, we introduce recon-all-clinical, a novel method for cortical reconstruction, registration, parcellation, and thickness estimation in brain MRI scans of any resolution and contrast. Our approach employs a hybrid analysis method that combines a convolutional neural network (CNN) trained with domain randomization to predict signed distance functions (SDFs) and classical geometry processing for accurate surface placement while maintaining topological and geometric constraints. The method does not require retraining for different acquisitions, thus simplifying the analysis of heterogeneous clinical datasets. We tested recon-all-clinical on multiple datasets, including over 19,000 clinical scans. The method consistently produced precise cortical reconstructions and high parcellation accuracy across varied MRI contrasts and resolutions. Cortical thickness estimates are precise enough to capture aging effects independently of MRI contrast, although accuracy varies with slice thickness. Our method is publicly available at https://surfer.nmr.mgh.harvard.edu/fswiki/recon-all-clinical, enabling researchers to perform detailed cortical analysis on the huge amounts of already existing clinical MRI scans. This advancement may be particularly valuable for studying rare diseases and underrepresented populations where research-grade MRI data is scarce.
Cortical analysis of heterogeneous clinical brain MRI scans for large-scale neuroimaging studies
May 02, 2023Abstract:Surface analysis of the cortex is ubiquitous in human neuroimaging with MRI, e.g., for cortical registration, parcellation, or thickness estimation. The convoluted cortical geometry requires isotropic scans (e.g., 1mm MPRAGEs) and good gray-white matter contrast for 3D reconstruction. This precludes the analysis of most brain MRI scans acquired for clinical purposes. Analyzing such scans would enable neuroimaging studies with sample sizes that cannot be achieved with current research datasets, particularly for underrepresented populations and rare diseases. Here we present the first method for cortical reconstruction, registration, parcellation, and thickness estimation for clinical brain MRI scans of any resolution and pulse sequence. The methods has a learning component and a classical optimization module. The former uses domain randomization to train a CNN that predicts an implicit representation of the white matter and pial surfaces (a signed distance function) at 1mm isotropic resolution, independently of the pulse sequence and resolution of the input. The latter uses geometry processing to place the surfaces while accurately satisfying topological and geometric constraints, thus enabling subsequent parcellation and thickness estimation with existing methods. We present results on 5mm axial FLAIR scans from ADNI and on a highly heterogeneous clinical dataset with 5,000 scans. Code and data are publicly available at https://surfer.nmr.mgh.harvard.edu/fswiki/recon-all-clinical
Anatomy-aware and acquisition-agnostic joint registration with SynthMorph
Jan 26, 2023Abstract:Affine image registration is a cornerstone of medical-image processing and analysis. While classical algorithms can achieve excellent accuracy, they solve a time-consuming optimization for every new image pair. Deep-learning (DL) methods learn a function that maps an image pair to an output transform. Evaluating the functions is fast, but capturing large transforms can be challenging, and networks tend to struggle if a test-image characteristic shifts from the training domain, such as the contrast or resolution. A majority of affine methods are also agnostic to the anatomy the user wishes to align; the registration will be inaccurate if algorithms consider all structures in the image. We address these shortcomings with a fast, robust, and easy-to-use DL tool for affine and deformable registration of any brain image without preprocessing, right off the MRI scanner. First, we rigorously analyze how competing architectures learn affine transforms across a diverse set of neuroimaging data, aiming to truly capture the behavior of methods in the real world. Second, we leverage a recent strategy to train networks with wildly varying images synthesized from label maps, yielding robust performance across acquisition specifics. Third, we optimize the spatial overlap of select anatomical labels, which enables networks to distinguish between anatomy of interest and irrelevant structures, removing the need for preprocessing that excludes content that would otherwise reduce the accuracy of anatomy-specific registration. We combine the affine model with prior work on deformable registration and test brain-specific registration across a landscape of MRI protocols unseen at training, demonstrating consistent and improved accuracy compared to existing tools. We distribute our code and tool at https://w3id.org/synthmorph, providing a single complete end-to-end solution for registration of brain MRI.
An Open-Source Tool for Longitudinal Whole-Brain and White Matter Lesion Segmentation
Jul 10, 2022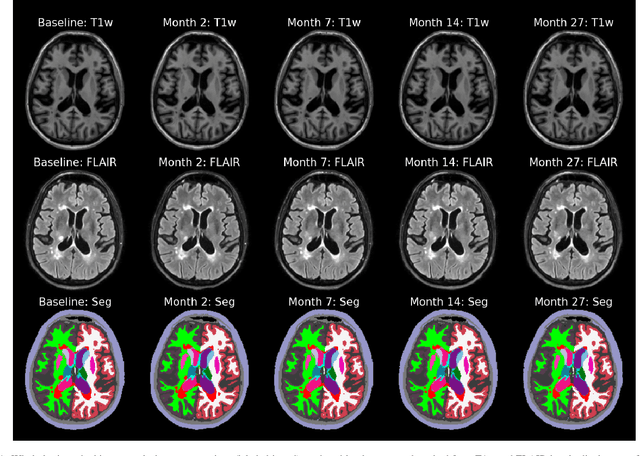
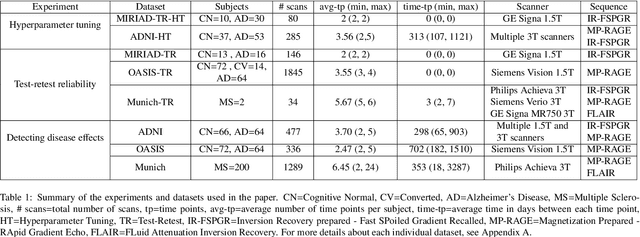
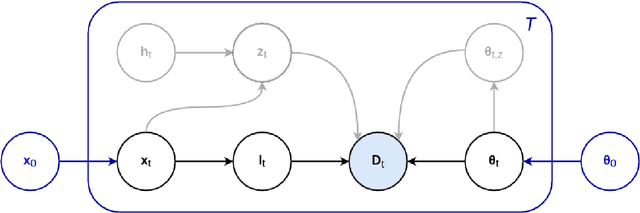
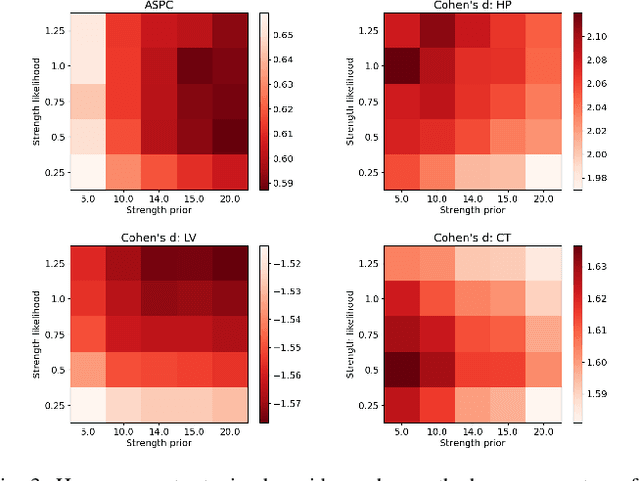
Abstract:In this paper we describe and validate a longitudinal method for whole-brain segmentation of longitudinal MRI scans. It builds upon an existing whole-brain segmentation method that can handle multi-contrast data and robustly analyze images with white matter lesions. This method is here extended with subject-specific latent variables that encourage temporal consistency between its segmentation results, enabling it to better track subtle morphological changes in dozens of neuroanatomical structures and white matter lesions. We validate the proposed method on multiple datasets of control subjects and patients suffering from Alzheimer's disease and multiple sclerosis, and compare its results against those obtained with its original cross-sectional formulation and two benchmark longitudinal methods. The results indicate that the method attains a higher test-retest reliability, while being more sensitive to longitudinal disease effect differences between patient groups. An implementation is publicly available as part of the open-source neuroimaging package FreeSurfer.
Learning the Effect of Registration Hyperparameters with HyperMorph
Mar 30, 2022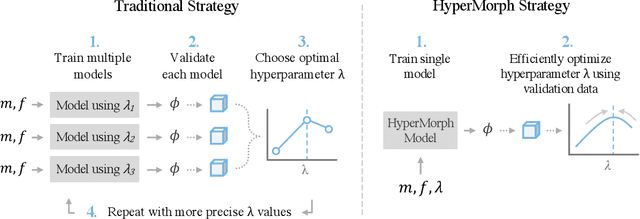
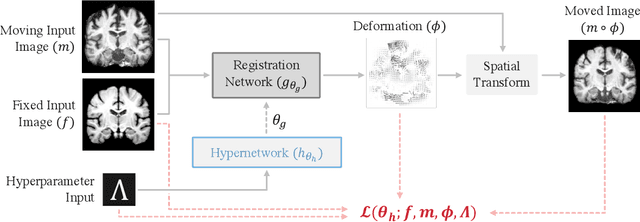

Abstract:We introduce HyperMorph, a framework that facilitates efficient hyperparameter tuning in learning-based deformable image registration. Classical registration algorithms perform an iterative pair-wise optimization to compute a deformation field that aligns two images. Recent learning-based approaches leverage large image datasets to learn a function that rapidly estimates a deformation for a given image pair. In both strategies, the accuracy of the resulting spatial correspondences is strongly influenced by the choice of certain hyperparameter values. However, an effective hyperparameter search consumes substantial time and human effort as it often involves training multiple models for different fixed hyperparameter values and may lead to suboptimal registration. We propose an amortized hyperparameter learning strategy to alleviate this burden by learning the impact of hyperparameters on deformation fields. We design a meta network, or hypernetwork, that predicts the parameters of a registration network for input hyperparameters, thereby comprising a single model that generates the optimal deformation field corresponding to given hyperparameter values. This strategy enables fast, high-resolution hyperparameter search at test-time, reducing the inefficiency of traditional approaches while increasing flexibility. We also demonstrate additional benefits of HyperMorph, including enhanced robustness to model initialization and the ability to rapidly identify optimal hyperparameter values specific to a dataset, image contrast, task, or even anatomical region, all without the need to retrain models. We make our code publicly available at http://hypermorph.voxelmorph.net.
SynthSeg: Domain Randomisation for Segmentation of Brain MRI Scans of any Contrast and Resolution
Jul 20, 2021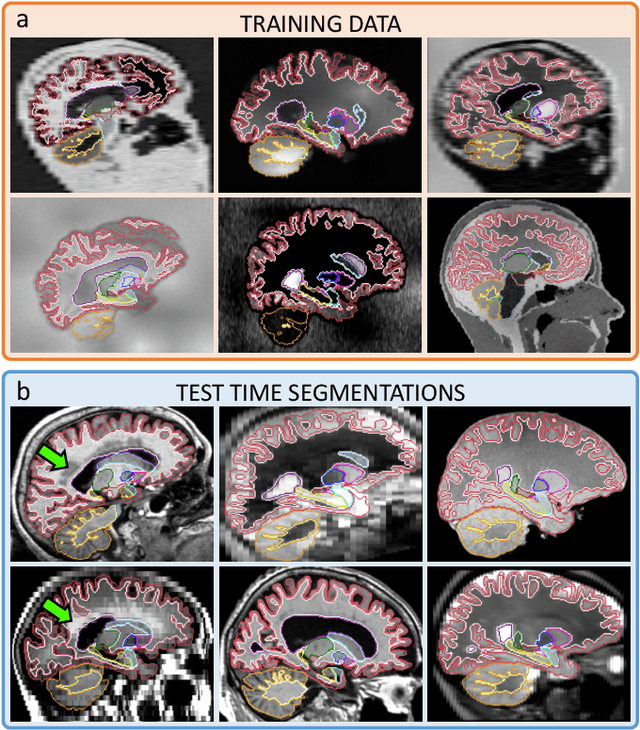
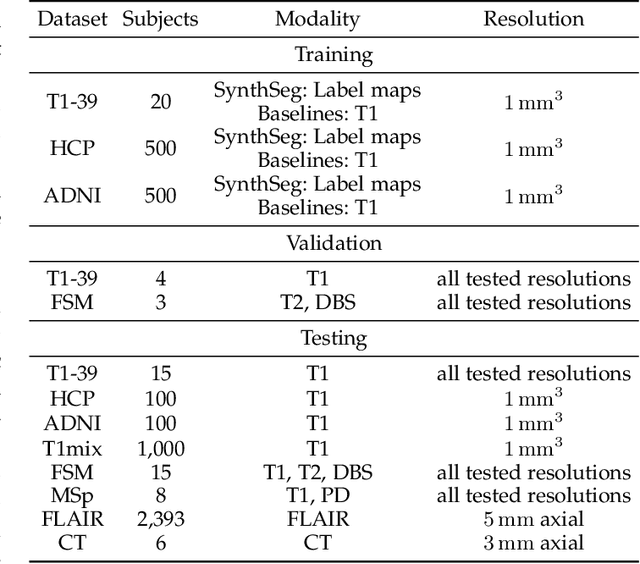
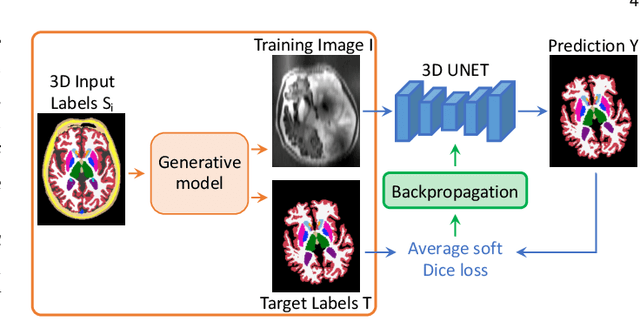
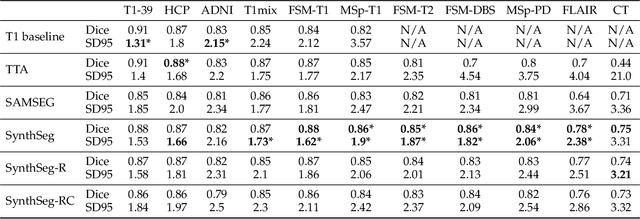
Abstract:Despite advances in data augmentation and transfer learning, convolutional neural networks (CNNs) have difficulties generalising to unseen target domains. When applied to segmentation of brain MRI scans, CNNs are highly sensitive to changes in resolution and contrast: even within the same MR modality, decreases in performance can be observed across datasets. We introduce SynthSeg, the first segmentation CNN agnostic to brain MRI scans of any contrast and resolution. SynthSeg is trained with synthetic data sampled from a generative model inspired by Bayesian segmentation. Crucially, we adopt a \textit{domain randomisation} strategy where we fully randomise the generation parameters to maximise the variability of the training data. Consequently, SynthSeg can segment preprocessed and unpreprocessed real scans of any target domain, without retraining or fine-tuning. Because SynthSeg only requires segmentations to be trained (no images), it can learn from label maps obtained automatically from existing datasets of different populations (e.g., with atrophy and lesions), thus achieving robustness to a wide range of morphological variability. We demonstrate SynthSeg on 5,500 scans of 6 modalities and 10 resolutions, where it exhibits unparalleled generalisation compared to supervised CNNs, test time adaptation, and Bayesian segmentation. The code and trained model are available at https://github.com/BBillot/SynthSeg.
A Longitudinal Method for Simultaneous Whole-Brain and Lesion Segmentation in Multiple Sclerosis
Sep 15, 2020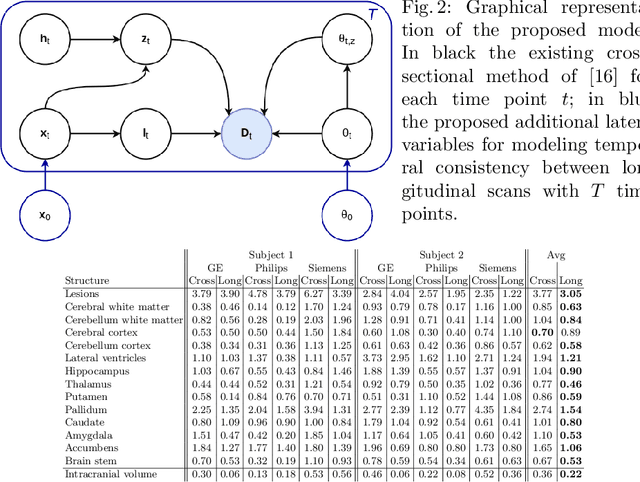
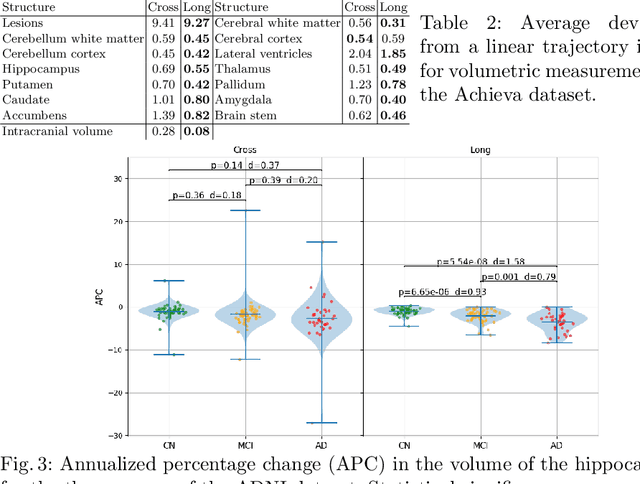
Abstract:In this paper we propose a novel method for the segmentation of longitudinal brain MRI scans of patients suffering from Multiple Sclerosis. The method builds upon an existing cross-sectional method for simultaneous whole-brain and lesion segmentation, introducing subject-specific latent variables to encourage temporal consistency between longitudinal scans. It is very generally applicable, as it does not make any prior assumptions on the scanner, the MRI protocol, or the number and timing of longitudinal follow-up scans. Preliminary experiments on three longitudinal datasets indicate that the proposed method produces more reliable segmentations and detects disease effects better than the cross-sectional method it is based upon.
Kinetic Compressive Sensing
Mar 27, 2018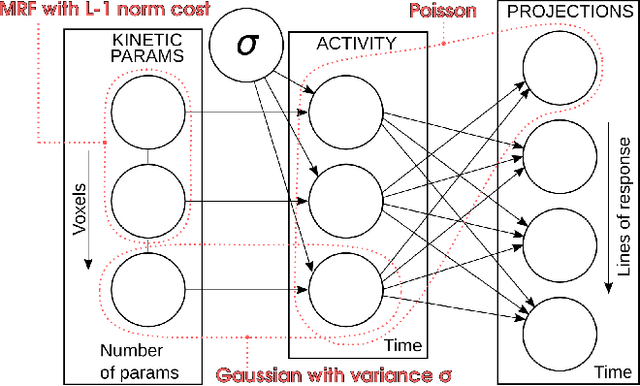
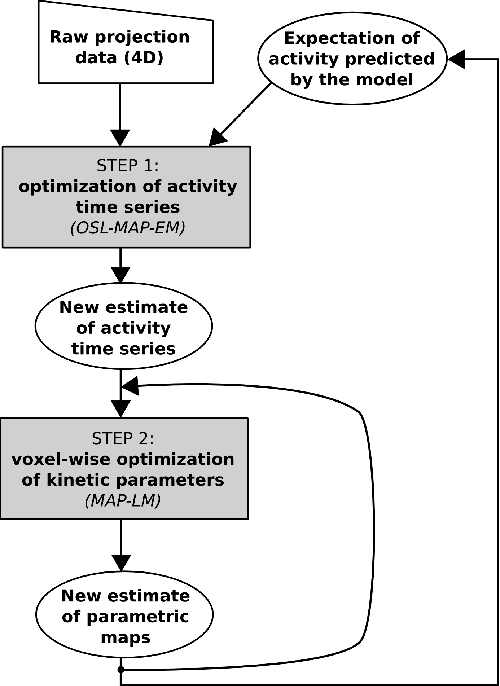

Abstract:Parametric images provide insight into the spatial distribution of physiological parameters, but they are often extremely noisy, due to low SNR of tomographic data. Direct estimation from projections allows accurate noise modeling, improving the results of post-reconstruction fitting. We propose a method, which we name kinetic compressive sensing (KCS), based on a hierarchical Bayesian model and on a novel reconstruction algorithm, that encodes sparsity of kinetic parameters. Parametric maps are reconstructed by maximizing the joint probability, with an Iterated Conditional Modes (ICM) approach, alternating the optimization of activity time series (OS-MAP-OSL), and kinetic parameters (MAP-LM). We evaluated the proposed algorithm on a simulated dynamic phantom: a bias/variance study confirmed how direct estimates can improve the quality of parametric maps over a post-reconstruction fitting, and showed how the novel sparsity prior can further reduce their variance, without affecting bias. Real FDG PET human brain data (Siemens mMR, 40min) images were also processed. Results enforced how the proposed KCS-regularized direct method can produce spatially coherent images and parametric maps, with lower spatial noise and better tissue contrast. A GPU-based open source implementation of the algorithm is provided.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge