Colin Magdamo
End-to-end Cortical Surface Reconstruction from Clinical Magnetic Resonance Images
May 20, 2025Abstract:Surface-based cortical analysis is valuable for a variety of neuroimaging tasks, such as spatial normalization, parcellation, and gray matter (GM) thickness estimation. However, most tools for estimating cortical surfaces work exclusively on scans with at least 1 mm isotropic resolution and are tuned to a specific magnetic resonance (MR) contrast, often T1-weighted (T1w). This precludes application using most clinical MR scans, which are very heterogeneous in terms of contrast and resolution. Here, we use synthetic domain-randomized data to train the first neural network for explicit estimation of cortical surfaces from scans of any contrast and resolution, without retraining. Our method deforms a template mesh to the white matter (WM) surface, which guarantees topological correctness. This mesh is further deformed to estimate the GM surface. We compare our method to recon-all-clinical (RAC), an implicit surface reconstruction method which is currently the only other tool capable of processing heterogeneous clinical MR scans, on ADNI and a large clinical dataset (n=1,332). We show a approximately 50 % reduction in cortical thickness error (from 0.50 to 0.24 mm) with respect to RAC and better recovery of the aging-related cortical thinning patterns detected by FreeSurfer on high-resolution T1w scans. Our method enables fast and accurate surface reconstruction of clinical scans, allowing studies (1) with sample sizes far beyond what is feasible in a research setting, and (2) of clinical populations that are difficult to enroll in research studies. The code is publicly available at https://github.com/simnibs/brainnet.
Recon-all-clinical: Cortical surface reconstruction and analysis of heterogeneous clinical brain MRI
Sep 05, 2024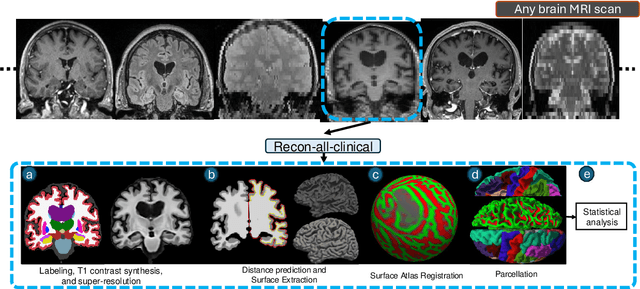
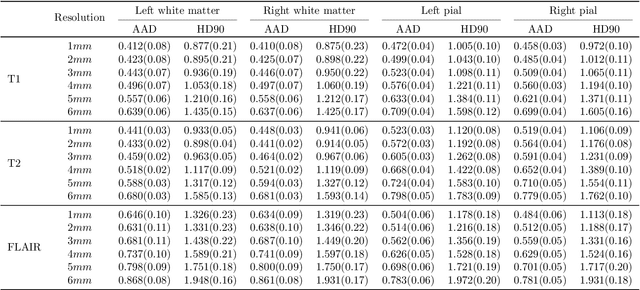
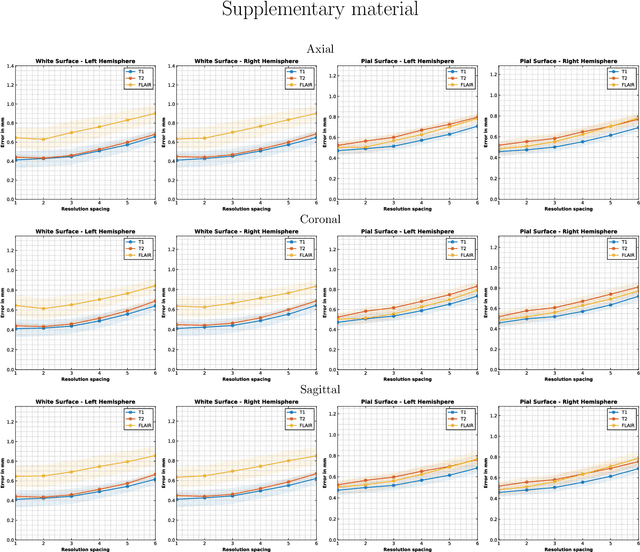
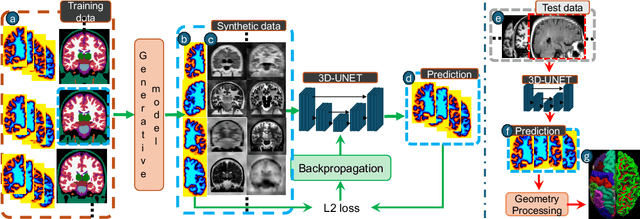
Abstract:Surface-based analysis of the cerebral cortex is ubiquitous in human neuroimaging with MRI. It is crucial for cortical registration, parcellation, and thickness estimation. Traditionally, these analyses require high-resolution, isotropic scans with good gray-white matter contrast, typically a 1mm T1-weighted scan. This excludes most clinical MRI scans, which are often anisotropic and lack the necessary T1 contrast. To enable large-scale neuroimaging studies using vast clinical data, we introduce recon-all-clinical, a novel method for cortical reconstruction, registration, parcellation, and thickness estimation in brain MRI scans of any resolution and contrast. Our approach employs a hybrid analysis method that combines a convolutional neural network (CNN) trained with domain randomization to predict signed distance functions (SDFs) and classical geometry processing for accurate surface placement while maintaining topological and geometric constraints. The method does not require retraining for different acquisitions, thus simplifying the analysis of heterogeneous clinical datasets. We tested recon-all-clinical on multiple datasets, including over 19,000 clinical scans. The method consistently produced precise cortical reconstructions and high parcellation accuracy across varied MRI contrasts and resolutions. Cortical thickness estimates are precise enough to capture aging effects independently of MRI contrast, although accuracy varies with slice thickness. Our method is publicly available at https://surfer.nmr.mgh.harvard.edu/fswiki/recon-all-clinical, enabling researchers to perform detailed cortical analysis on the huge amounts of already existing clinical MRI scans. This advancement may be particularly valuable for studying rare diseases and underrepresented populations where research-grade MRI data is scarce.
Leveraging Pre-trained and Transformer-derived Embeddings from EHRs to Characterize Heterogeneity Across Alzheimer's Disease and Related Dementias
Mar 30, 2024



Abstract:Alzheimer's disease is a progressive, debilitating neurodegenerative disease that affects 50 million people globally. Despite this substantial health burden, available treatments for the disease are limited and its fundamental causes remain poorly understood. Previous work has suggested the existence of clinically-meaningful sub-types, which it is suggested may correspond to distinct etiologies, disease courses, and ultimately appropriate treatments. Here, we use unsupervised learning techniques on electronic health records (EHRs) from a cohort of memory disorder patients to characterise heterogeneity in this disease population. Pre-trained embeddings for medical codes as well as transformer-derived Clinical BERT embeddings of free text are used to encode patient EHRs. We identify the existence of sub-populations on the basis of comorbidities and shared textual features, and discuss their clinical significance.
Cortical analysis of heterogeneous clinical brain MRI scans for large-scale neuroimaging studies
May 02, 2023Abstract:Surface analysis of the cortex is ubiquitous in human neuroimaging with MRI, e.g., for cortical registration, parcellation, or thickness estimation. The convoluted cortical geometry requires isotropic scans (e.g., 1mm MPRAGEs) and good gray-white matter contrast for 3D reconstruction. This precludes the analysis of most brain MRI scans acquired for clinical purposes. Analyzing such scans would enable neuroimaging studies with sample sizes that cannot be achieved with current research datasets, particularly for underrepresented populations and rare diseases. Here we present the first method for cortical reconstruction, registration, parcellation, and thickness estimation for clinical brain MRI scans of any resolution and pulse sequence. The methods has a learning component and a classical optimization module. The former uses domain randomization to train a CNN that predicts an implicit representation of the white matter and pial surfaces (a signed distance function) at 1mm isotropic resolution, independently of the pulse sequence and resolution of the input. The latter uses geometry processing to place the surfaces while accurately satisfying topological and geometric constraints, thus enabling subsequent parcellation and thickness estimation with existing methods. We present results on 5mm axial FLAIR scans from ADNI and on a highly heterogeneous clinical dataset with 5,000 scans. Code and data are publicly available at https://surfer.nmr.mgh.harvard.edu/fswiki/recon-all-clinical
Robust machine learning segmentation for large-scale analysis of heterogeneous clinical brain MRI datasets
Sep 05, 2022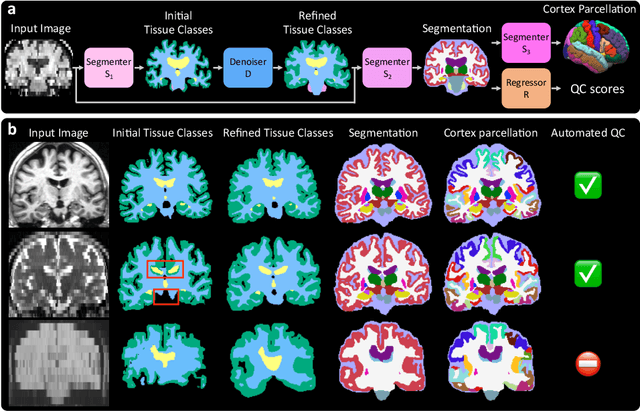
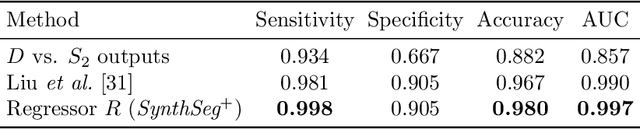
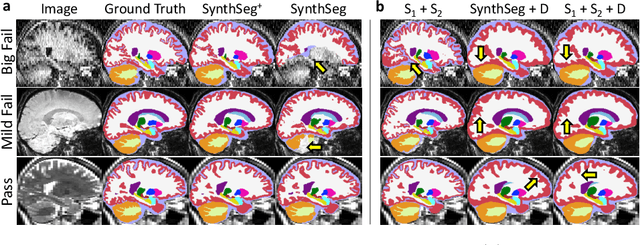

Abstract:Every year, millions of brain MRI scans are acquired in hospitals, which is a figure considerably larger than the size of any research dataset. Therefore, the ability to analyse such scans could transform neuroimaging research. Yet, their potential remains untapped, since no automated algorithm can cope with the high variability in clinical acquisitions (MR contrast, resolution, orientation, etc.). Here we present SynthSeg+, an AI segmentation suite that enables, for the first time, robust analysis of heterogeneous clinical datasets. Specifically, in addition to whole-brain segmentation, SynthSeg+ also performs cortical parcellation, intracranial volume estimation, and automated detection of faulty segmentations (mainly caused by scans of very low quality). We demonstrate SynthSeg+ in seven experiments, including an ageing study on 14,000 scans, where it accurately replicates atrophy patterns observed on data of much higher quality. SynthSeg+ is publicly released as a ready-to-use tool to unlock the potential of quantitative morphometry in wide-ranging settings.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge