Sudeshna Das
Massachusetts General Hospital, Boston, MA
Graph AI generates neurological hypotheses validated in molecular, organoid, and clinical systems
Dec 13, 2025Abstract:Neurological diseases are the leading global cause of disability, yet most lack disease-modifying treatments. We present PROTON, a heterogeneous graph transformer that generates testable hypotheses across molecular, organoid, and clinical systems. To evaluate PROTON, we apply it to Parkinson's disease (PD), bipolar disorder (BD), and Alzheimer's disease (AD). In PD, PROTON linked genetic risk loci to genes essential for dopaminergic neuron survival and predicted pesticides toxic to patient-derived neurons, including the insecticide endosulfan, which ranked within the top 1.29% of predictions. In silico screens performed by PROTON reproduced six genome-wide $α$-synuclein experiments, including a split-ubiquitin yeast two-hybrid system (normalized enrichment score [NES] = 2.30, FDR-adjusted $p < 1 \times 10^{-4}$), an ascorbate peroxidase proximity labeling assay (NES = 2.16, FDR $< 1 \times 10^{-4}$), and a high-depth targeted exome sequencing study in 496 synucleinopathy patients (NES = 2.13, FDR $< 1 \times 10^{-4}$). In BD, PROTON predicted calcitriol as a candidate drug that reversed proteomic alterations observed in cortical organoids derived from BD patients. In AD, we evaluated PROTON predictions in health records from $n = 610,524$ patients at Mass General Brigham, confirming that five PROTON-predicted drugs were associated with reduced seven-year dementia risk (minimum hazard ratio = 0.63, 95% CI: 0.53-0.75, $p < 1 \times 10^{-7}$). PROTON generated neurological hypotheses that were evaluated across molecular, organoid, and clinical systems, defining a path for AI-driven discovery in neurological disease.
Identifying social isolation themes in NVDRS text narratives using topic modeling and text-classification methods
Jun 18, 2025Abstract:Social isolation and loneliness, which have been increasing in recent years strongly contribute toward suicide rates. Although social isolation and loneliness are not currently recorded within the US National Violent Death Reporting System's (NVDRS) structured variables, natural language processing (NLP) techniques can be used to identify these constructs in law enforcement and coroner medical examiner narratives. Using topic modeling to generate lexicon development and supervised learning classifiers, we developed high-quality classifiers (average F1: .86, accuracy: .82). Evaluating over 300,000 suicides from 2002 to 2020, we identified 1,198 mentioning chronic social isolation. Decedents had higher odds of chronic social isolation classification if they were men (OR = 1.44; CI: 1.24, 1.69, p<.0001), gay (OR = 3.68; 1.97, 6.33, p<.0001), or were divorced (OR = 3.34; 2.68, 4.19, p<.0001). We found significant predictors for other social isolation topics of recent or impending divorce, child custody loss, eviction or recent move, and break-up. Our methods can improve surveillance and prevention of social isolation and loneliness in the United States.
End-to-end Cortical Surface Reconstruction from Clinical Magnetic Resonance Images
May 20, 2025Abstract:Surface-based cortical analysis is valuable for a variety of neuroimaging tasks, such as spatial normalization, parcellation, and gray matter (GM) thickness estimation. However, most tools for estimating cortical surfaces work exclusively on scans with at least 1 mm isotropic resolution and are tuned to a specific magnetic resonance (MR) contrast, often T1-weighted (T1w). This precludes application using most clinical MR scans, which are very heterogeneous in terms of contrast and resolution. Here, we use synthetic domain-randomized data to train the first neural network for explicit estimation of cortical surfaces from scans of any contrast and resolution, without retraining. Our method deforms a template mesh to the white matter (WM) surface, which guarantees topological correctness. This mesh is further deformed to estimate the GM surface. We compare our method to recon-all-clinical (RAC), an implicit surface reconstruction method which is currently the only other tool capable of processing heterogeneous clinical MR scans, on ADNI and a large clinical dataset (n=1,332). We show a approximately 50 % reduction in cortical thickness error (from 0.50 to 0.24 mm) with respect to RAC and better recovery of the aging-related cortical thinning patterns detected by FreeSurfer on high-resolution T1w scans. Our method enables fast and accurate surface reconstruction of clinical scans, allowing studies (1) with sample sizes far beyond what is feasible in a research setting, and (2) of clinical populations that are difficult to enroll in research studies. The code is publicly available at https://github.com/simnibs/brainnet.
HILGEN: Hierarchically-Informed Data Generation for Biomedical NER Using Knowledgebases and Large Language Models
Mar 06, 2025Abstract:We present HILGEN, a Hierarchically-Informed Data Generation approach that combines domain knowledge from the Unified Medical Language System (UMLS) with synthetic data generated by large language models (LLMs), specifically GPT-3.5. Our approach leverages UMLS's hierarchical structure to expand training data with related concepts, while incorporating contextual information from LLMs through targeted prompts aimed at automatically generating synthetic examples for sparsely occurring named entities. The performance of the HILGEN approach was evaluated across four biomedical NER datasets (MIMIC III, BC5CDR, NCBI-Disease, and Med-Mentions) using BERT-Large and DANN (Data Augmentation with Nearest Neighbor Classifier) models, applying various data generation strategies, including UMLS, GPT-3.5, and their best ensemble. For the BERT-Large model, incorporating UMLS led to an average F1 score improvement of 40.36%, while using GPT-3.5 resulted in a comparable average increase of 40.52%. The Best-Ensemble approach using BERT-Large achieved the highest improvement, with an average increase of 42.29%. DANN model's F1 score improved by 22.74% on average using the UMLS-only approach. The GPT-3.5-based method resulted in a 21.53% increase, and the Best-Ensemble DANN model showed a more notable improvement, with an average increase of 25.03%. Our proposed HILGEN approach improves NER performance in few-shot settings without requiring additional manually annotated data. Our experiments demonstrate that an effective strategy for optimizing biomedical NER is to combine biomedical knowledge curated in the past, such as the UMLS, and generative LLMs to create synthetic training instances. Our future research will focus on exploring additional innovative synthetic data generation strategies for further improving NER performance.
Multi Scale Graph Neural Network for Alzheimer's Disease
Nov 16, 2024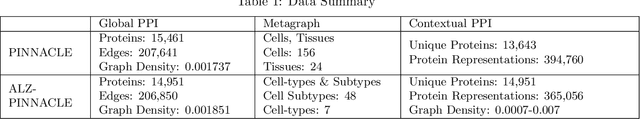
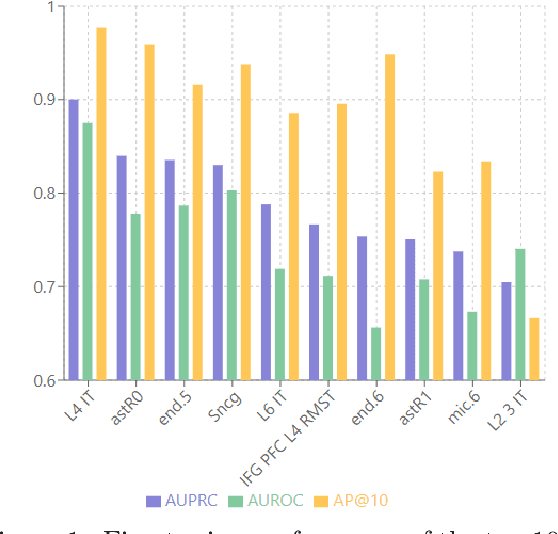
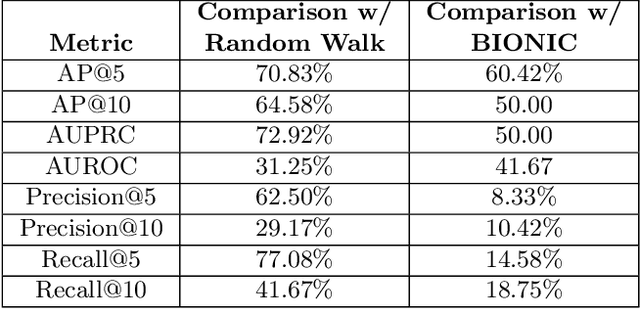
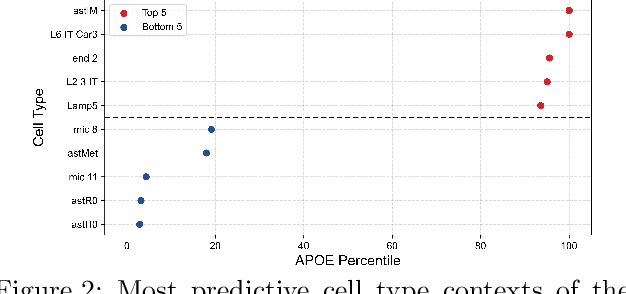
Abstract:Alzheimer's disease (AD) is a complex, progressive neurodegenerative disorder characterized by extracellular A\b{eta} plaques, neurofibrillary tau tangles, glial activation, and neuronal degeneration, involving multiple cell types and pathways. Current models often overlook the cellular context of these pathways. To address this, we developed a multiscale graph neural network (GNN) model, ALZ PINNACLE, using brain omics data from donors spanning the entire aging to AD spectrum. ALZ PINNACLE is based on the PINNACLE GNN framework, which learns context-aware protein, cell type, and tissue representations within a unified latent space. ALZ PINNACLE was trained on 14,951 proteins, 206,850 protein interactions, 7 cell types, and 48 cell subtypes or states. After pretraining, we investigated the learned embedding of APOE, the largest genetic risk factor for AD, across different cell types. Notably, APOE embeddings showed high similarity in microglial, neuronal, and CD8 cells, suggesting a similar role of APOE in these cell types. Fine tuning the model on AD risk genes revealed cell type contexts predictive of the role of APOE in AD. Our results suggest that ALZ PINNACLE may provide a valuable framework for uncovering novel insights into AD neurobiology.
Recon-all-clinical: Cortical surface reconstruction and analysis of heterogeneous clinical brain MRI
Sep 05, 2024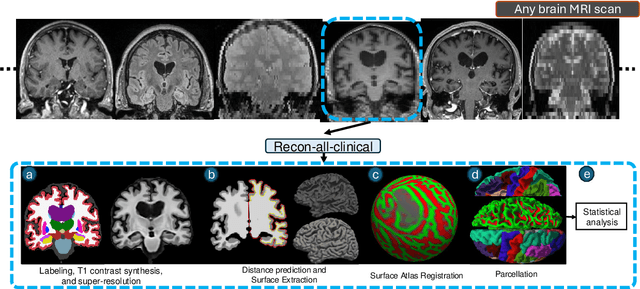
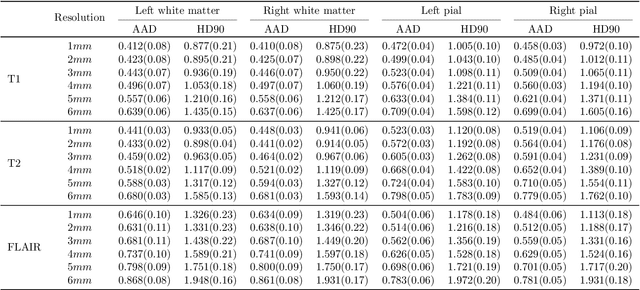
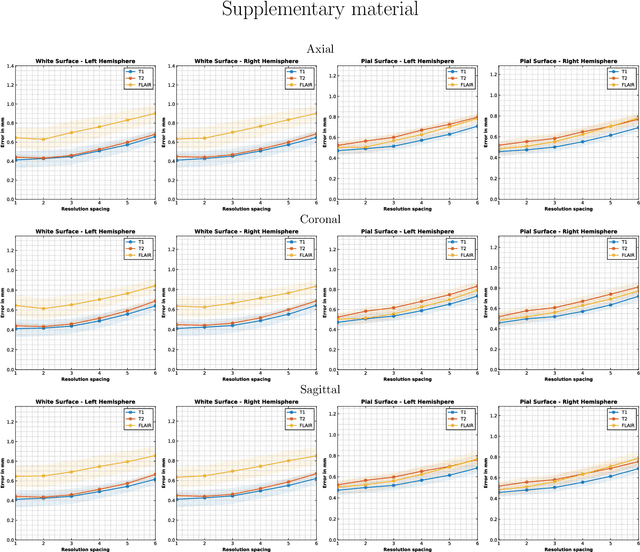
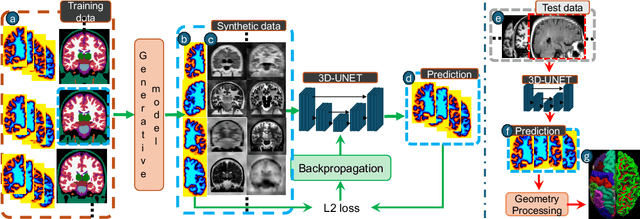
Abstract:Surface-based analysis of the cerebral cortex is ubiquitous in human neuroimaging with MRI. It is crucial for cortical registration, parcellation, and thickness estimation. Traditionally, these analyses require high-resolution, isotropic scans with good gray-white matter contrast, typically a 1mm T1-weighted scan. This excludes most clinical MRI scans, which are often anisotropic and lack the necessary T1 contrast. To enable large-scale neuroimaging studies using vast clinical data, we introduce recon-all-clinical, a novel method for cortical reconstruction, registration, parcellation, and thickness estimation in brain MRI scans of any resolution and contrast. Our approach employs a hybrid analysis method that combines a convolutional neural network (CNN) trained with domain randomization to predict signed distance functions (SDFs) and classical geometry processing for accurate surface placement while maintaining topological and geometric constraints. The method does not require retraining for different acquisitions, thus simplifying the analysis of heterogeneous clinical datasets. We tested recon-all-clinical on multiple datasets, including over 19,000 clinical scans. The method consistently produced precise cortical reconstructions and high parcellation accuracy across varied MRI contrasts and resolutions. Cortical thickness estimates are precise enough to capture aging effects independently of MRI contrast, although accuracy varies with slice thickness. Our method is publicly available at https://surfer.nmr.mgh.harvard.edu/fswiki/recon-all-clinical, enabling researchers to perform detailed cortical analysis on the huge amounts of already existing clinical MRI scans. This advancement may be particularly valuable for studying rare diseases and underrepresented populations where research-grade MRI data is scarce.
Two-layer retrieval augmented generation framework for low-resource medical question-answering: proof of concept using Reddit data
May 29, 2024



Abstract:Retrieval augmented generation (RAG) provides the capability to constrain generative model outputs, and mitigate the possibility of hallucination, by providing relevant in-context text. The number of tokens a generative large language model (LLM) can incorporate as context is finite, thus limiting the volume of knowledge from which to generate an answer. We propose a two-layer RAG framework for query-focused answer generation and evaluate a proof-of-concept for this framework in the context of query-focused summary generation from social media forums, focusing on emerging drug-related information. The evaluations demonstrate the effectiveness of the two-layer framework in resource constrained settings to enable researchers in obtaining near real-time data from users.
Reddit-Impacts: A Named Entity Recognition Dataset for Analyzing Clinical and Social Effects of Substance Use Derived from Social Media
May 09, 2024



Abstract:Substance use disorders (SUDs) are a growing concern globally, necessitating enhanced understanding of the problem and its trends through data-driven research. Social media are unique and important sources of information about SUDs, particularly since the data in such sources are often generated by people with lived experiences. In this paper, we introduce Reddit-Impacts, a challenging Named Entity Recognition (NER) dataset curated from subreddits dedicated to discussions on prescription and illicit opioids, as well as medications for opioid use disorder. The dataset specifically concentrates on the lesser-studied, yet critically important, aspects of substance use--its clinical and social impacts. We collected data from chosen subreddits using the publicly available Application Programming Interface for Reddit. We manually annotated text spans representing clinical and social impacts reported by people who also reported personal nonmedical use of substances including but not limited to opioids, stimulants and benzodiazepines. Our objective is to create a resource that can enable the development of systems that can automatically detect clinical and social impacts of substance use from text-based social media data. The successful development of such systems may enable us to better understand how nonmedical use of substances affects individual health and societal dynamics, aiding the development of effective public health strategies. In addition to creating the annotated data set, we applied several machine learning models to establish baseline performances. Specifically, we experimented with transformer models like BERT, and RoBERTa, one few-shot learning model DANN by leveraging the full training dataset, and GPT-3.5 by using one-shot learning, for automatic NER of clinical and social impacts. The dataset has been made available through the 2024 SMM4H shared tasks.
CARE-SD: Classifier-based analysis for recognizing and eliminating stigmatizing and doubt marker labels in electronic health records: model development and validation
May 08, 2024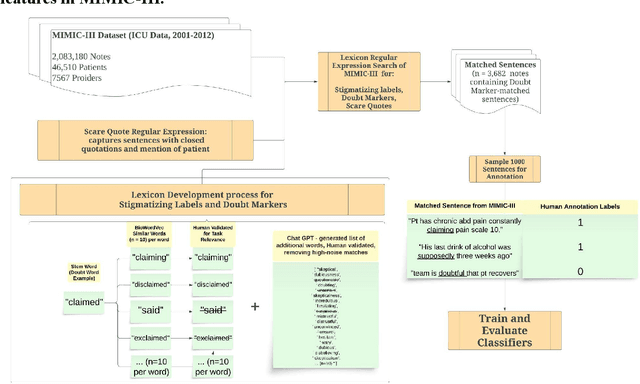
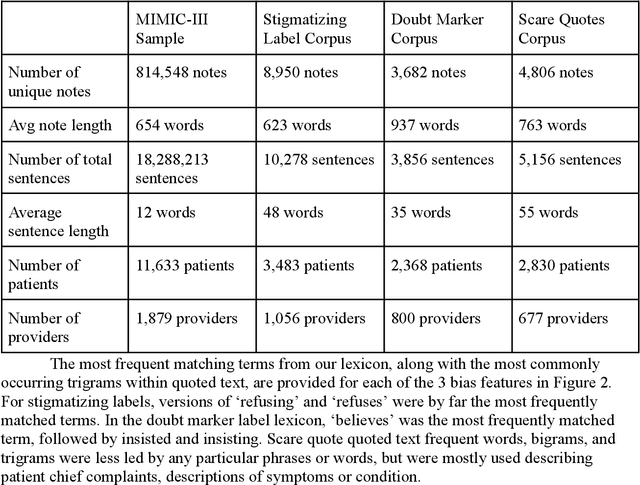
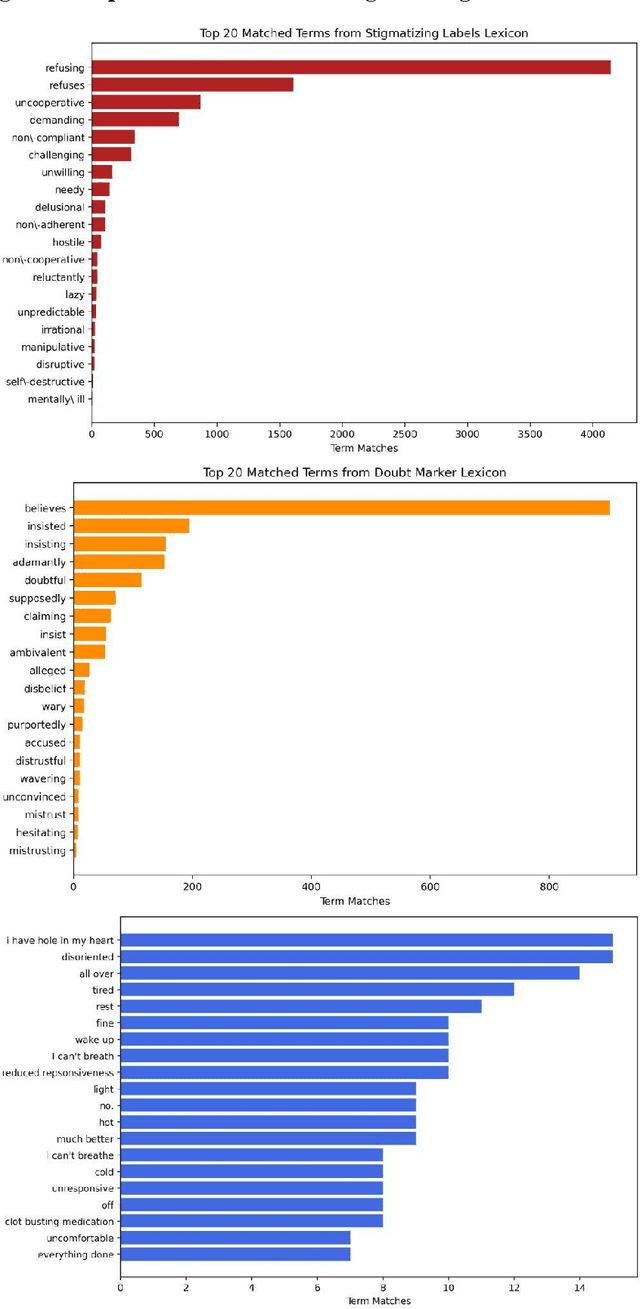
Abstract:Objective: To detect and classify features of stigmatizing and biased language in intensive care electronic health records (EHRs) using natural language processing techniques. Materials and Methods: We first created a lexicon and regular expression lists from literature-driven stem words for linguistic features of stigmatizing patient labels, doubt markers, and scare quotes within EHRs. The lexicon was further extended using Word2Vec and GPT 3.5, and refined through human evaluation. These lexicons were used to search for matches across 18 million sentences from the de-identified Medical Information Mart for Intensive Care-III (MIMIC-III) dataset. For each linguistic bias feature, 1000 sentence matches were sampled, labeled by expert clinical and public health annotators, and used to supervised learning classifiers. Results: Lexicon development from expanded literature stem-word lists resulted in a doubt marker lexicon containing 58 expressions, and a stigmatizing labels lexicon containing 127 expressions. Classifiers for doubt markers and stigmatizing labels had the highest performance, with macro F1-scores of .84 and .79, positive-label recall and precision values ranging from .71 to .86, and accuracies aligning closely with human annotator agreement (.87). Discussion: This study demonstrated the feasibility of supervised classifiers in automatically identifying stigmatizing labels and doubt markers in medical text, and identified trends in stigmatizing language use in an EHR setting. Additional labeled data may help improve lower scare quote model performance. Conclusions: Classifiers developed in this study showed high model performance and can be applied to identify patterns and target interventions to reduce stigmatizing labels and doubt markers in healthcare systems.
Leveraging Pre-trained and Transformer-derived Embeddings from EHRs to Characterize Heterogeneity Across Alzheimer's Disease and Related Dementias
Mar 30, 2024



Abstract:Alzheimer's disease is a progressive, debilitating neurodegenerative disease that affects 50 million people globally. Despite this substantial health burden, available treatments for the disease are limited and its fundamental causes remain poorly understood. Previous work has suggested the existence of clinically-meaningful sub-types, which it is suggested may correspond to distinct etiologies, disease courses, and ultimately appropriate treatments. Here, we use unsupervised learning techniques on electronic health records (EHRs) from a cohort of memory disorder patients to characterise heterogeneity in this disease population. Pre-trained embeddings for medical codes as well as transformer-derived Clinical BERT embeddings of free text are used to encode patient EHRs. We identify the existence of sub-populations on the basis of comorbidities and shared textual features, and discuss their clinical significance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge