Graciela Gonzalez-Hernandez
Reddit-Impacts: A Named Entity Recognition Dataset for Analyzing Clinical and Social Effects of Substance Use Derived from Social Media
May 09, 2024



Abstract:Substance use disorders (SUDs) are a growing concern globally, necessitating enhanced understanding of the problem and its trends through data-driven research. Social media are unique and important sources of information about SUDs, particularly since the data in such sources are often generated by people with lived experiences. In this paper, we introduce Reddit-Impacts, a challenging Named Entity Recognition (NER) dataset curated from subreddits dedicated to discussions on prescription and illicit opioids, as well as medications for opioid use disorder. The dataset specifically concentrates on the lesser-studied, yet critically important, aspects of substance use--its clinical and social impacts. We collected data from chosen subreddits using the publicly available Application Programming Interface for Reddit. We manually annotated text spans representing clinical and social impacts reported by people who also reported personal nonmedical use of substances including but not limited to opioids, stimulants and benzodiazepines. Our objective is to create a resource that can enable the development of systems that can automatically detect clinical and social impacts of substance use from text-based social media data. The successful development of such systems may enable us to better understand how nonmedical use of substances affects individual health and societal dynamics, aiding the development of effective public health strategies. In addition to creating the annotated data set, we applied several machine learning models to establish baseline performances. Specifically, we experimented with transformer models like BERT, and RoBERTa, one few-shot learning model DANN by leveraging the full training dataset, and GPT-3.5 by using one-shot learning, for automatic NER of clinical and social impacts. The dataset has been made available through the 2024 SMM4H shared tasks.
ReportAGE: Automatically extracting the exact age of Twitter users based on self-reports in tweets
Mar 10, 2021


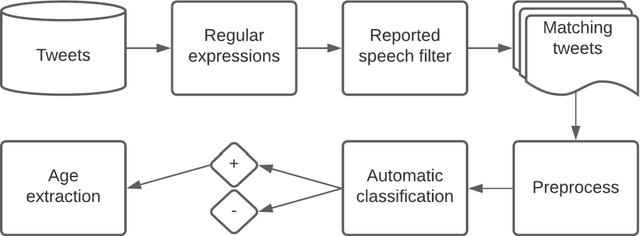
Abstract:Advancing the utility of social media data for research applications requires methods for automatically detecting demographic information about social media study populations, including users' age. The objective of this study was to develop and evaluate a method that automatically identifies the exact age of users based on self-reports in their tweets. Our end-to-end automatic natural language processing (NLP) pipeline, ReportAGE, includes query patterns to retrieve tweets that potentially mention an age, a classifier to distinguish retrieved tweets that self-report the user's exact age ("age" tweets) and those that do not ("no age" tweets), and rule-based extraction to identify the age. To develop and evaluate ReportAGE, we manually annotated 11,000 tweets that matched the query patterns. Based on 1000 tweets that were annotated by all five annotators, inter-annotator agreement (Fleiss' kappa) was 0.80 for distinguishing "age" and "no age" tweets, and 0.95 for identifying the exact age among the "age" tweets on which the annotators agreed. A deep neural network classifier, based on a RoBERTa-Large pretrained model, achieved the highest F1-score of 0.914 (precision = 0.905, recall = 0.942) for the "age" class. When the age extraction was evaluated using the classifier's predictions, it achieved an F1-score of 0.855 (precision = 0.805, recall = 0.914) for the "age" class. When it was evaluated directly on the held-out test set, it achieved an F1-score of 0.931 (precision = 0.873, recall = 0.998) for the "age" class. We deployed ReportAGE on more than 1.2 billion tweets posted by 245,927 users, and predicted ages for 132,637 (54%) of them. Scaling the detection of exact age to this large number of users can advance the utility of social media data for research applications that do not align with the predefined age groupings of extant binary or multi-class classification approaches.
Towards Automatic Bot Detection in Twitter for Health-related Tasks
Sep 29, 2019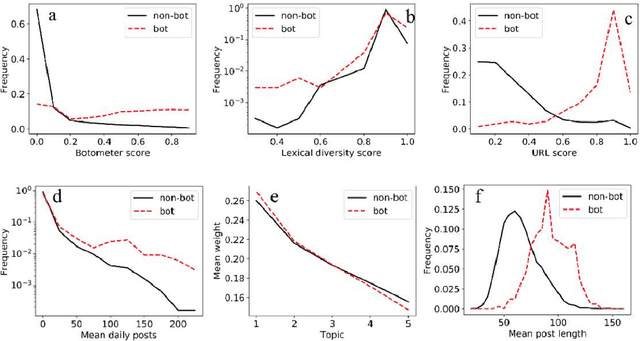

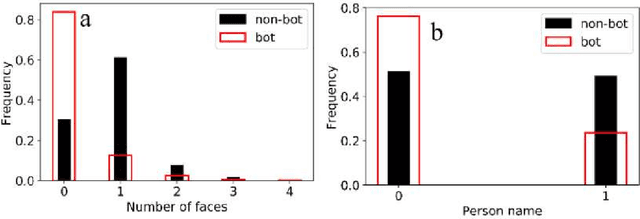
Abstract:With the increasing use of social media data for health-related research, the credibility of the information from this source has been questioned as the posts may originate from automated accounts or "bots". While automatic bot detection approaches have been proposed, there are none that have been evaluated on users posting health-related information. In this paper, we extend an existing bot detection system and customize it for health-related research. Using a dataset of Twitter users, we first show that the system, which was designed for political bot detection, underperforms when applied to health-related Twitter users. We then incorporate additional features and a statistical machine learning classifier to significantly improve bot detection performance. Our approach obtains F_1 scores of 0.7 for the "bot" class, representing improvements of 0.339. Our approach is customizable and generalizable for bot detection in other health-related social media cohorts.
Automatically Identifying Comparator Groups on Twitter for Digital Epidemiology of Pregnancy Outcomes
Aug 16, 2019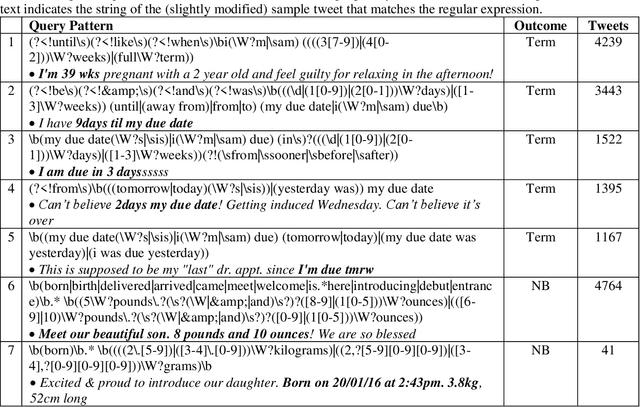
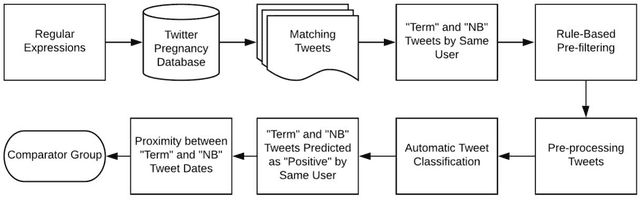

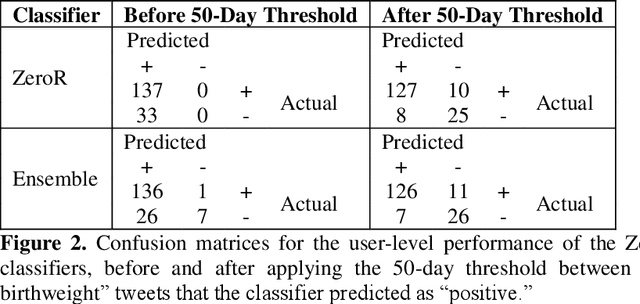
Abstract:Despite the prevalence of adverse pregnancy outcomes such as miscarriage, stillbirth, birth defects, and preterm birth, their causes are largely unknown. We seek to advance the use of social media for observational studies of pregnancy outcomes by developing a natural language processing pipeline for automatically identifying users from which to select comparator groups on Twitter. We annotated 2361 tweets by users who have announced their pregnancy on Twitter, which were used to train and evaluate supervised machine learning algorithms as a basis for automatically detecting women who have reported that their pregnancy had reached term and their baby was born at a normal weight. Upon further processing the tweet-level predictions of a majority voting-based ensemble classifier, the pipeline achieved a user-level F1-score of 0.933, with a precision of 0.947 and a recall of 0.920. Our pipeline will be deployed to identify large comparator groups for studying pregnancy outcomes on Twitter.
Deep Neural Networks Ensemble for Detecting Medication Mentions in Tweets
Apr 10, 2019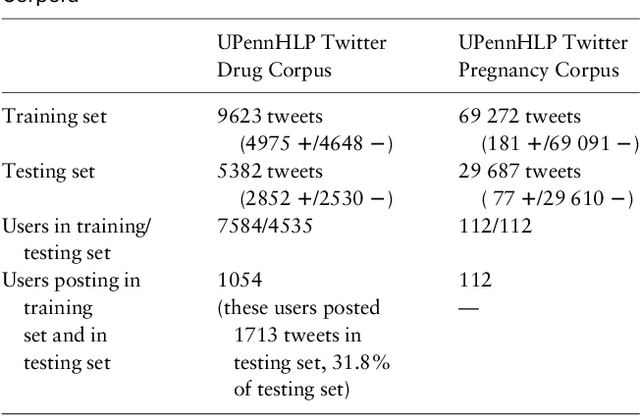


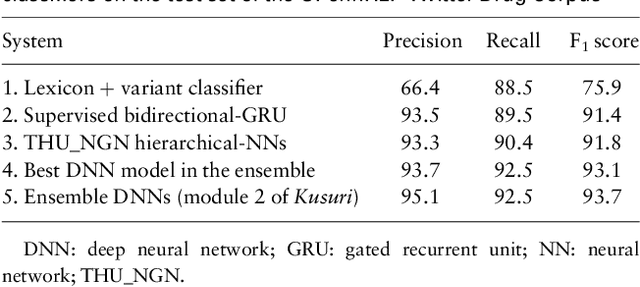
Abstract:Objective: After years of research, Twitter posts are now recognized as an important source of patient-generated data, providing unique insights into population health. A fundamental step to incorporating Twitter data in pharmacoepidemiological research is to automatically recognize medication mentions in tweets. Given that lexical searches for medication names may fail due to misspellings or ambiguity with common words, we propose a more advanced method to recognize them. Methods: We present Kusuri, an Ensemble Learning classifier, able to identify tweets mentioning drug products and dietary supplements. Kusuri ("medication" in Japanese) is composed of two modules. First, four different classifiers (lexicon-based, spelling-variant-based, pattern-based and one based on a weakly-trained neural network) are applied in parallel to discover tweets potentially containing medication names. Second, an ensemble of deep neural networks encoding morphological, semantical and long-range dependencies of important words in the tweets discovered is used to make the final decision. Results: On a balanced (50-50) corpus of 15,005 tweets, Kusuri demonstrated performances close to human annotators with 93.7% F1-score, the best score achieved thus far on this corpus. On a corpus made of all tweets posted by 113 Twitter users (98,959 tweets, with only 0.26% mentioning medications), Kusuri obtained 76.3% F1-score. There is not a prior drug extraction system that compares running on such an extremely unbalanced dataset. Conclusion: The system identifies tweets mentioning drug names with performance high enough to ensure its usefulness and ready to be integrated in larger natural language processing systems.
Automatically Detecting Self-Reported Birth Defect Outcomes on Twitter for Large-scale Epidemiological Research
Oct 22, 2018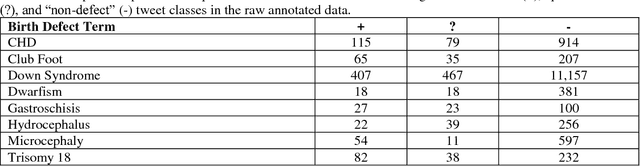
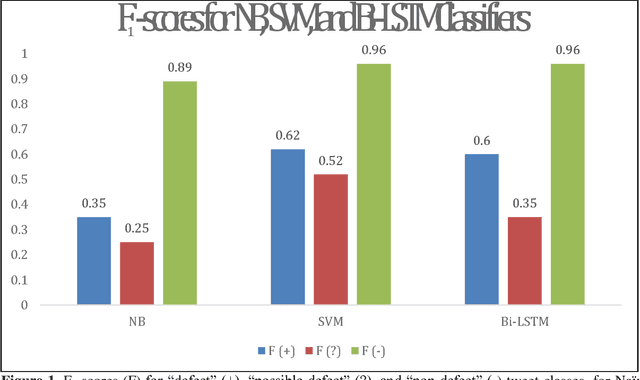
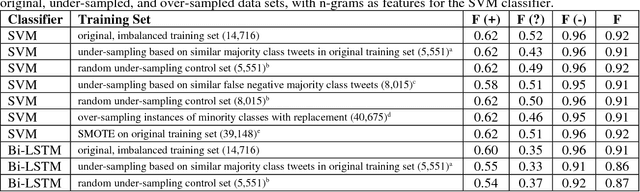
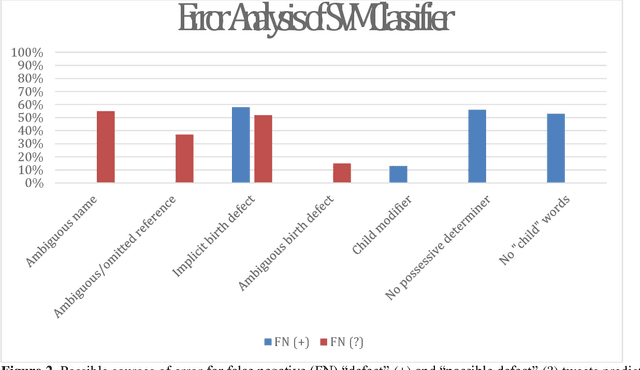
Abstract:In recent work, we identified and studied a small cohort of Twitter users whose pregnancies with birth defect outcomes could be observed via their publicly available tweets. Exploiting social media's large-scale potential to complement the limited methods for studying birth defects, the leading cause of infant mortality, depends on the further development of automatic methods. The primary objective of this study was to take the first step towards scaling the use of social media for observing pregnancies with birth defect outcomes, namely, developing methods for automatically detecting tweets by users reporting their birth defect outcomes. We annotated and pre-processed approximately 23,000 tweets that mention birth defects in order to train and evaluate supervised machine learning algorithms, including feature-engineered and deep learning-based classifiers. We also experimented with various under-sampling and over-sampling approaches to address the class imbalance. A Support Vector Machine (SVM) classifier trained on the original, imbalanced data set, with n-grams, word clusters, and structural features, achieved the best baseline performance for the positive classes: an F1-score of 0.65 for the "defect" class and 0.51 for the "possible defect" class. Our contributions include (i) natural language processing (NLP) and supervised machine learning methods for automatically detecting tweets by users reporting their birth defect outcomes, (ii) a comparison of feature-engineered and deep learning-based classifiers trained on imbalanced, under-sampled, and over-sampled data, and (iii) an error analysis that could inform classification improvements using our publicly available corpus. Future work will focus on automating user-level analyses for cohort inclusion.
An unsupervised and customizable misspelling generator for mining noisy health-related text sources
Jun 04, 2018

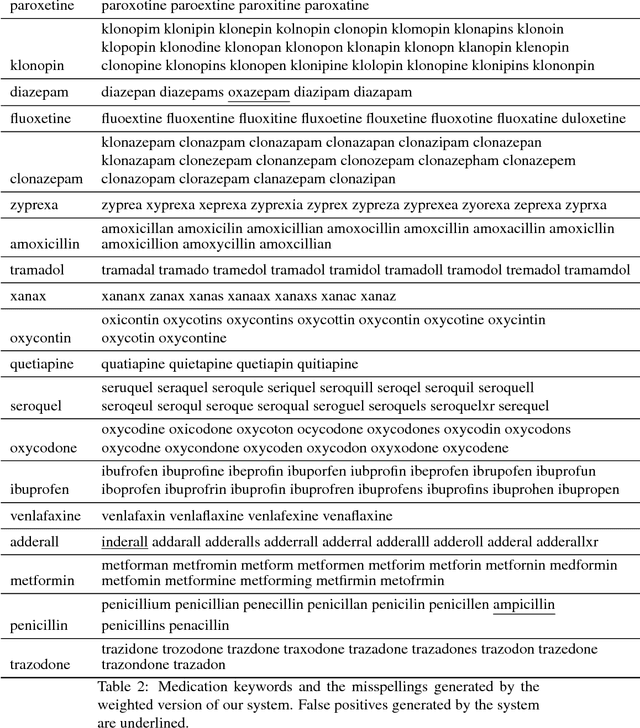

Abstract:In this paper, we present a customizable datacentric system that automatically generates common misspellings for complex health-related terms. The spelling variant generator relies on a dense vector model learned from large unlabeled text, which is used to find semantically close terms to the original/seed keyword, followed by the filtering of terms that are lexically dissimilar beyond a given threshold. The process is executed recursively, converging when no new terms similar (lexically and semantically) to the seed keyword are found. Weighting of intra-word character sequence similarities allows further problem-specific customization of the system. On a dataset prepared for this study, our system outperforms the current state-of-the-art for medication name variant generation with best F1-score of 0.69 and F1/4-score of 0.78. Extrinsic evaluation of the system on a set of cancer-related terms showed an increase of over 67% in retrieval rate from Twitter posts when the generated variants are included. Our proposed spelling variant generator has several advantages over the current state-of-the-art and other types of variant generators-(i) it is capable of filtering out lexically similar but semantically dissimilar terms, (ii) the number of variants generated is low as many low-frequency and ambiguous misspellings are filtered out, and (iii) the system is fully automatic, customizable and easily executable. While the base system is fully unsupervised, we show how supervision maybe employed to adjust weights for task-specific customization. The performance and significant relative simplicity of our proposed approach makes it a much needed misspelling generation resource for health-related text mining from noisy sources. The source code for the system has been made publicly available for research purposes.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge