Davy Weissenbacher
LIPN
Deep Neural Networks Ensemble for Detecting Medication Mentions in Tweets
Apr 10, 2019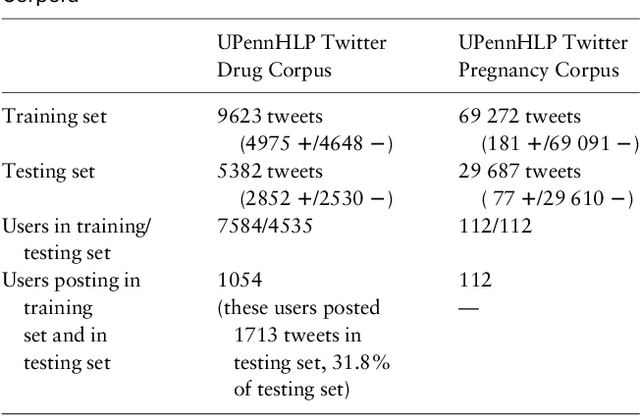


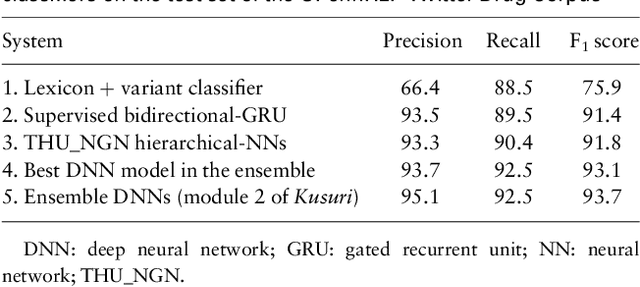
Abstract:Objective: After years of research, Twitter posts are now recognized as an important source of patient-generated data, providing unique insights into population health. A fundamental step to incorporating Twitter data in pharmacoepidemiological research is to automatically recognize medication mentions in tweets. Given that lexical searches for medication names may fail due to misspellings or ambiguity with common words, we propose a more advanced method to recognize them. Methods: We present Kusuri, an Ensemble Learning classifier, able to identify tweets mentioning drug products and dietary supplements. Kusuri ("medication" in Japanese) is composed of two modules. First, four different classifiers (lexicon-based, spelling-variant-based, pattern-based and one based on a weakly-trained neural network) are applied in parallel to discover tweets potentially containing medication names. Second, an ensemble of deep neural networks encoding morphological, semantical and long-range dependencies of important words in the tweets discovered is used to make the final decision. Results: On a balanced (50-50) corpus of 15,005 tweets, Kusuri demonstrated performances close to human annotators with 93.7% F1-score, the best score achieved thus far on this corpus. On a corpus made of all tweets posted by 113 Twitter users (98,959 tweets, with only 0.26% mentioning medications), Kusuri obtained 76.3% F1-score. There is not a prior drug extraction system that compares running on such an extremely unbalanced dataset. Conclusion: The system identifies tweets mentioning drug names with performance high enough to ensure its usefulness and ready to be integrated in larger natural language processing systems.
Automatically Detecting Self-Reported Birth Defect Outcomes on Twitter for Large-scale Epidemiological Research
Oct 22, 2018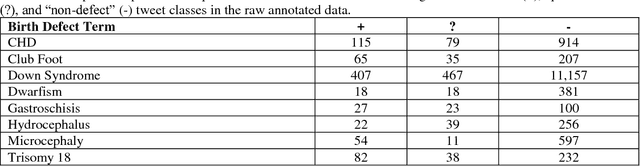
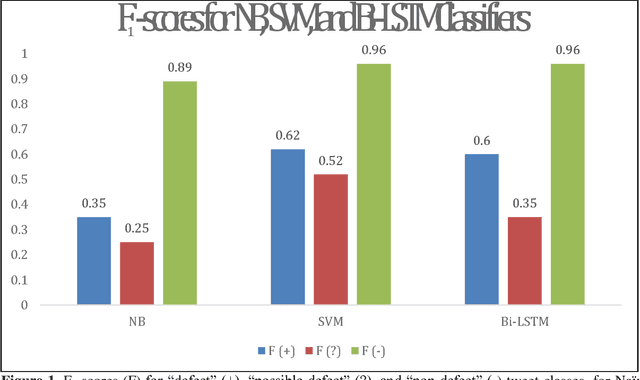
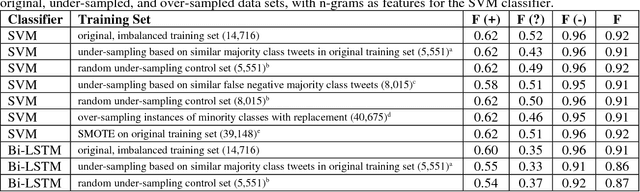
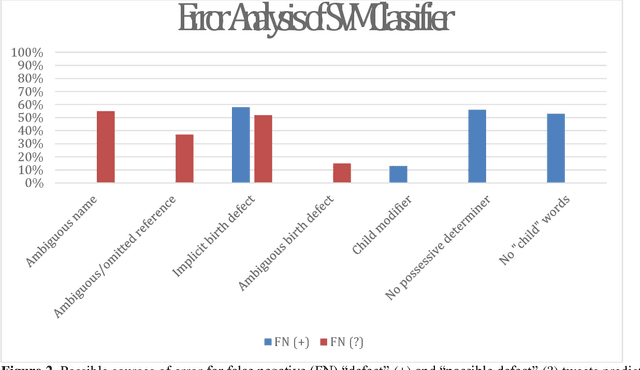
Abstract:In recent work, we identified and studied a small cohort of Twitter users whose pregnancies with birth defect outcomes could be observed via their publicly available tweets. Exploiting social media's large-scale potential to complement the limited methods for studying birth defects, the leading cause of infant mortality, depends on the further development of automatic methods. The primary objective of this study was to take the first step towards scaling the use of social media for observing pregnancies with birth defect outcomes, namely, developing methods for automatically detecting tweets by users reporting their birth defect outcomes. We annotated and pre-processed approximately 23,000 tweets that mention birth defects in order to train and evaluate supervised machine learning algorithms, including feature-engineered and deep learning-based classifiers. We also experimented with various under-sampling and over-sampling approaches to address the class imbalance. A Support Vector Machine (SVM) classifier trained on the original, imbalanced data set, with n-grams, word clusters, and structural features, achieved the best baseline performance for the positive classes: an F1-score of 0.65 for the "defect" class and 0.51 for the "possible defect" class. Our contributions include (i) natural language processing (NLP) and supervised machine learning methods for automatically detecting tweets by users reporting their birth defect outcomes, (ii) a comparison of feature-engineered and deep learning-based classifiers trained on imbalanced, under-sampled, and over-sampled data, and (iii) an error analysis that could inform classification improvements using our publicly available corpus. Future work will focus on automating user-level analyses for cohort inclusion.
The ALVIS Format for Linguistically Annotated Documents
Sep 24, 2006
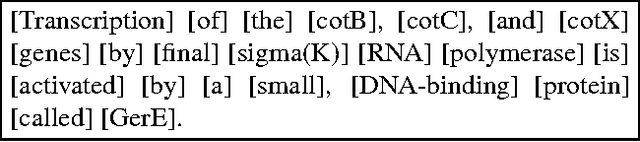
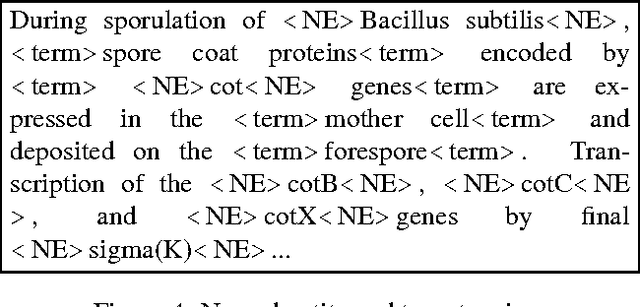

Abstract:The paper describes the ALVIS annotation format designed for the indexing of large collections of documents in topic-specific search engines. This paper is exemplified on the biological domain and on MedLine abstracts, as developing a specialized search engine for biologists is one of the ALVIS case studies. The ALVIS principle for linguistic annotations is based on existing works and standard propositions. We made the choice of stand-off annotations rather than inserted mark-up. Annotations are encoded as XML elements which form the linguistic subsection of the document record.
Event-based Information Extraction for the biomedical domain: the Caderige project
Sep 24, 2006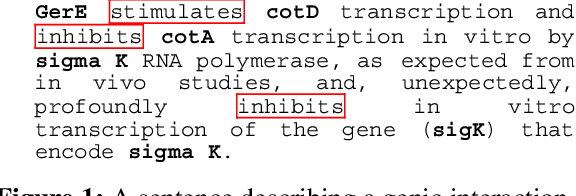
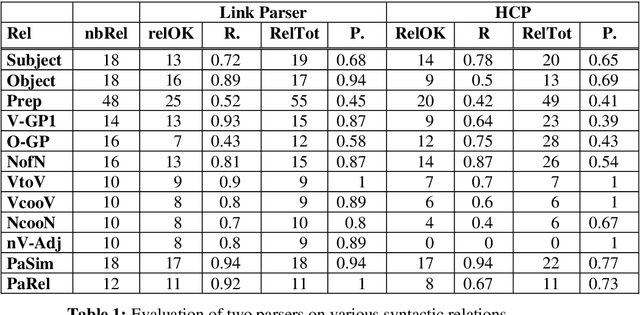
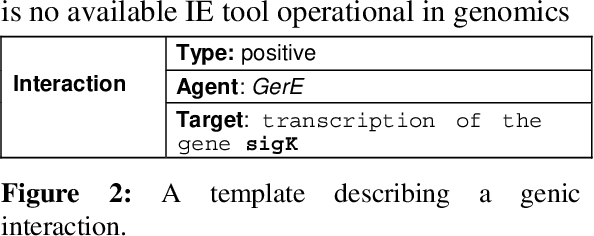
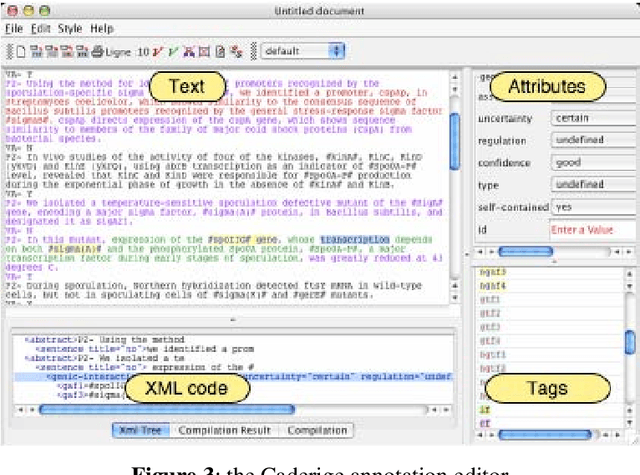
Abstract:This paper gives an overview of the Caderige project. This project involves teams from different areas (biology, machine learning, natural language processing) in order to develop high-level analysis tools for extracting structured information from biological bibliographical databases, especially Medline. The paper gives an overview of the approach and compares it to the state of the art.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge