Adeline Nazarenko
LIPN
A Benchmark Corpus for the Detection of Automatically Generated Text in Academic Publications
Feb 04, 2022
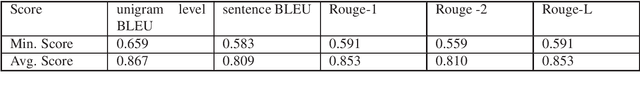
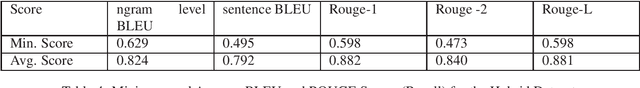

Abstract:Automatic text generation based on neural language models has achieved performance levels that make the generated text almost indistinguishable from those written by humans. Despite the value that text generation can have in various applications, it can also be employed for malicious tasks. The diffusion of such practices represent a threat to the quality of academic publishing. To address these problems, we propose in this paper two datasets comprised of artificially generated research content: a completely synthetic dataset and a partial text substitution dataset. In the first case, the content is completely generated by the GPT-2 model after a short prompt extracted from original papers. The partial or hybrid dataset is created by replacing several sentences of abstracts with sentences that are generated by the Arxiv-NLP model. We evaluate the quality of the datasets comparing the generated texts to aligned original texts using fluency metrics such as BLEU and ROUGE. The more natural the artificial texts seem, the more difficult they are to detect and the better is the benchmark. We also evaluate the difficulty of the task of distinguishing original from generated text by using state-of-the-art classification models.
A Robust Linguistic Platform for Efficient and Domain specific Web Content Analysis
Jun 29, 2007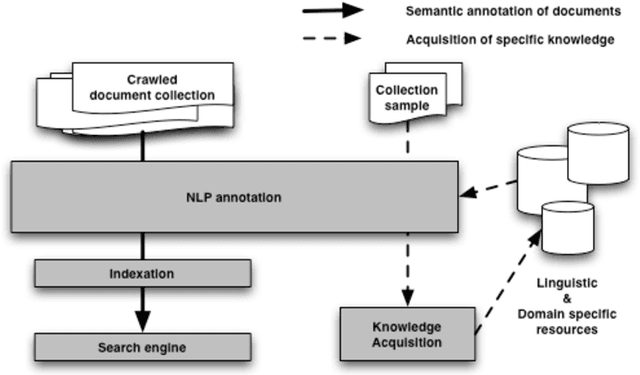
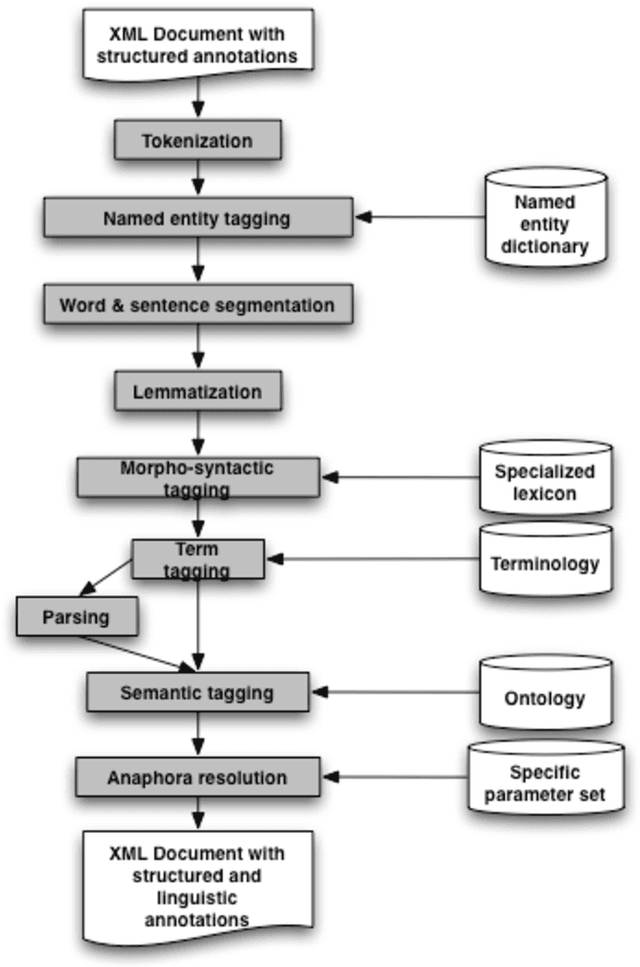

Abstract:Web semantic access in specific domains calls for specialized search engines with enhanced semantic querying and indexing capacities, which pertain both to information retrieval (IR) and to information extraction (IE). A rich linguistic analysis is required either to identify the relevant semantic units to index and weight them according to linguistic specific statistical distribution, or as the basis of an information extraction process. Recent developments make Natural Language Processing (NLP) techniques reliable enough to process large collections of documents and to enrich them with semantic annotations. This paper focuses on the design and the development of a text processing platform, Ogmios, which has been developed in the ALVIS project. The Ogmios platform exploits existing NLP modules and resources, which may be tuned to specific domains and produces linguistically annotated documents. We show how the three constraints of genericity, domain semantic awareness and performance can be handled all together.
Ontologies and Information Extraction
Sep 24, 2006



Abstract:This report argues that, even in the simplest cases, IE is an ontology-driven process. It is not a mere text filtering method based on simple pattern matching and keywords, because the extracted pieces of texts are interpreted with respect to a predefined partial domain model. This report shows that depending on the nature and the depth of the interpretation to be done for extracting the information, more or less knowledge must be involved. This report is mainly illustrated in biology, a domain in which there are critical needs for content-based exploration of the scientific literature and which becomes a major application domain for IE.
The ALVIS Format for Linguistically Annotated Documents
Sep 24, 2006
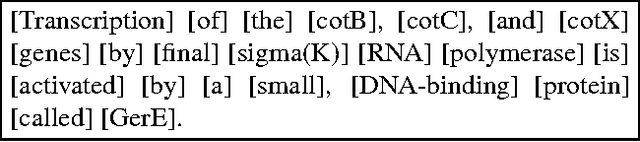
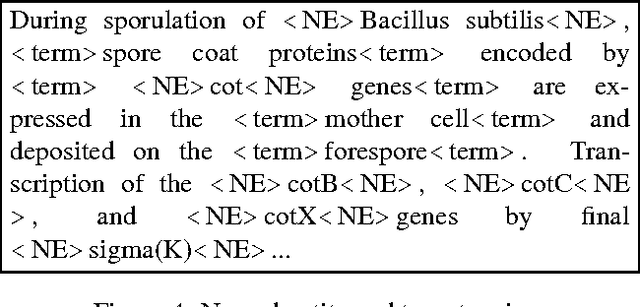

Abstract:The paper describes the ALVIS annotation format designed for the indexing of large collections of documents in topic-specific search engines. This paper is exemplified on the biological domain and on MedLine abstracts, as developing a specialized search engine for biologists is one of the ALVIS case studies. The ALVIS principle for linguistic annotations is based on existing works and standard propositions. We made the choice of stand-off annotations rather than inserted mark-up. Annotations are encoded as XML elements which form the linguistic subsection of the document record.
Event-based Information Extraction for the biomedical domain: the Caderige project
Sep 24, 2006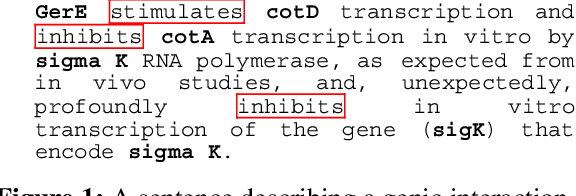
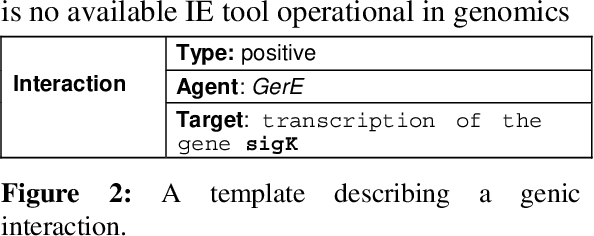
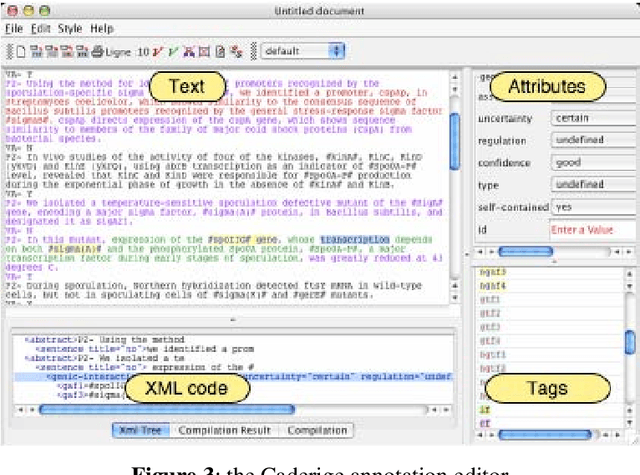
Abstract:This paper gives an overview of the Caderige project. This project involves teams from different areas (biology, machine learning, natural language processing) in order to develop high-level analysis tools for extracting structured information from biological bibliographical databases, especially Medline. The paper gives an overview of the approach and compares it to the state of the art.
Using NLP to build the hypertextuel network of a back-of-the-book index
Sep 24, 2006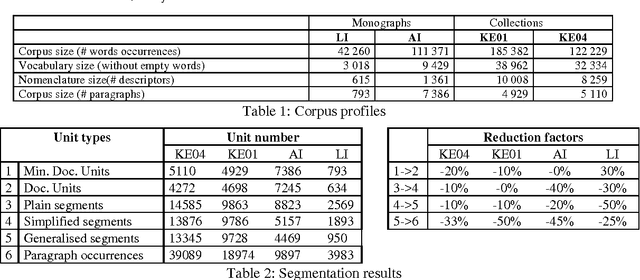
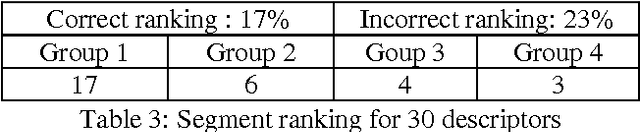
Abstract:Relying on the idea that back-of-the-book indexes are traditional devices for navigation through large documents, we have developed a method to build a hypertextual network that helps the navigation in a document. Building such an hypertextual network requires selecting a list of descriptors, identifying the relevant text segments to associate with each descriptor and finally ranking the descriptors and reference segments by relevance order. We propose a specific document segmentation method and a relevance measure for information ranking. The algorithms are tested on 4 corpora (of different types and domains) without human intervention or any semantic knowledge.
An application-oriented terminology evaluation: the case of back-of-the book indexes
Sep 24, 2006
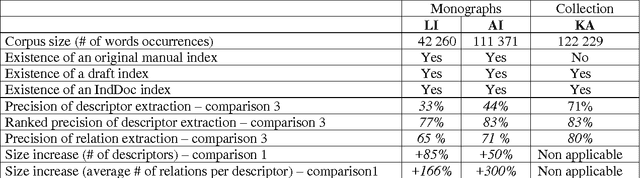
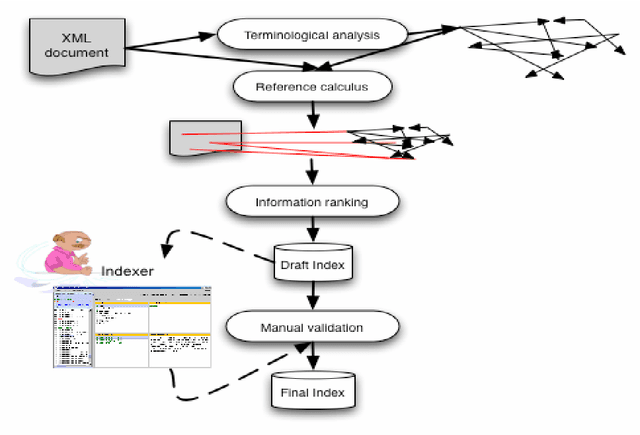
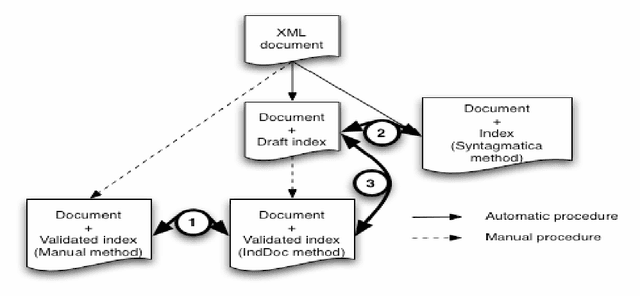
Abstract:This paper addresses the problem of computational terminology evaluation not per se but in a specific application context. This paper describes the evaluation procedure that has been used to assess the validity of our overall indexing approach and the quality of the IndDoc indexing tool. Even if user-oriented extended evaluation is irreplaceable, we argue that early evaluations are possible and they are useful for development guidance.
* 4 pages
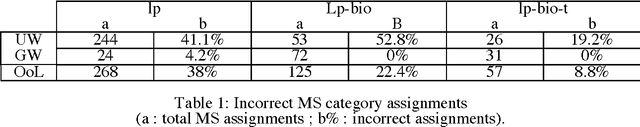
Lexical Adaptation of Link Grammar to the Biomedical Sublanguage: a Comparative Evaluation of Three Approaches
Jun 28, 2006



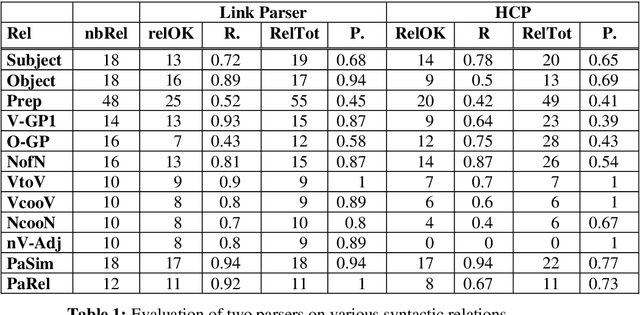
Abstract:We study the adaptation of Link Grammar Parser to the biomedical sublanguage with a focus on domain terms not found in a general parser lexicon. Using two biomedical corpora, we implement and evaluate three approaches to addressing unknown words: automatic lexicon expansion, the use of morphological clues, and disambiguation using a part-of-speech tagger. We evaluate each approach separately for its effect on parsing performance and consider combinations of these approaches. In addition to a 45% increase in parsing efficiency, we find that the best approach, incorporating information from a domain part-of-speech tagger, offers a statistically signicant 10% relative decrease in error. The adapted parser is available under an open-source license at http://www.it.utu.fi/biolg.
Adapting a general parser to a sublanguage
Jun 28, 2006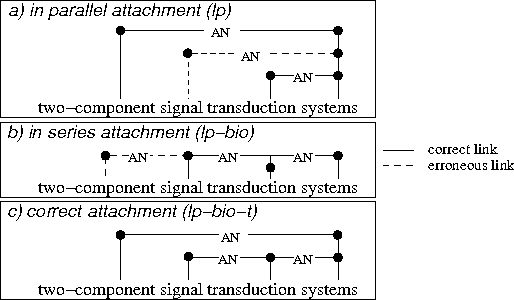
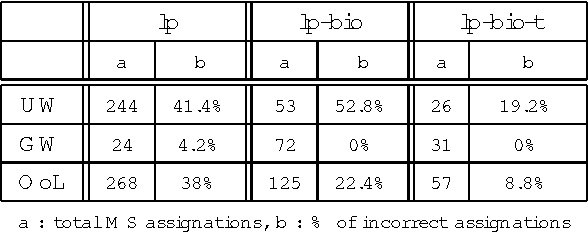
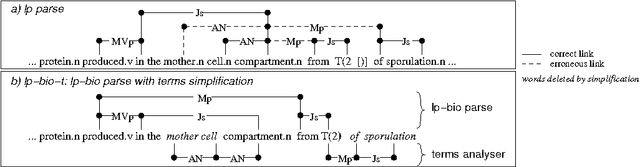
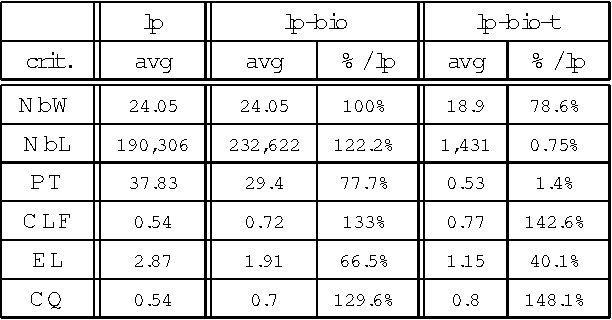
Abstract:In this paper, we propose a method to adapt a general parser (Link Parser) to sublanguages, focusing on the parsing of texts in biology. Our main proposal is the use of terminology (identication and analysis of terms) in order to reduce the complexity of the text to be parsed. Several other strategies are explored and finally combined among which text normalization, lexicon and morpho-guessing module extensions and grammar rules adaptation. We compare the parsing results before and after these adaptations.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge