Thierry Hamon
LIPN
A Robust Linguistic Platform for Efficient and Domain specific Web Content Analysis
Jun 29, 2007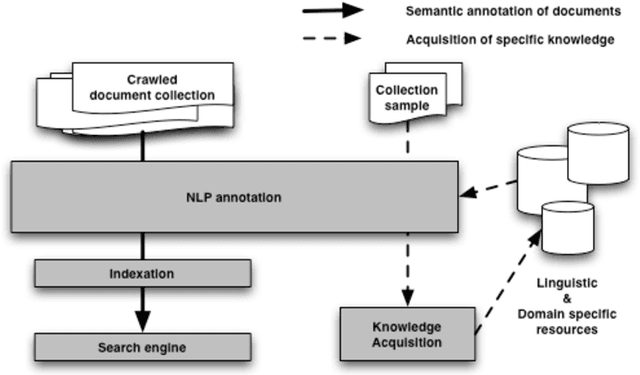

Abstract:Web semantic access in specific domains calls for specialized search engines with enhanced semantic querying and indexing capacities, which pertain both to information retrieval (IR) and to information extraction (IE). A rich linguistic analysis is required either to identify the relevant semantic units to index and weight them according to linguistic specific statistical distribution, or as the basis of an information extraction process. Recent developments make Natural Language Processing (NLP) techniques reliable enough to process large collections of documents and to enrich them with semantic annotations. This paper focuses on the design and the development of a text processing platform, Ogmios, which has been developed in the ALVIS project. The Ogmios platform exploits existing NLP modules and resources, which may be tuned to specific domains and produces linguistically annotated documents. We show how the three constraints of genericity, domain semantic awareness and performance can be handled all together.
The ALVIS Format for Linguistically Annotated Documents
Sep 24, 2006
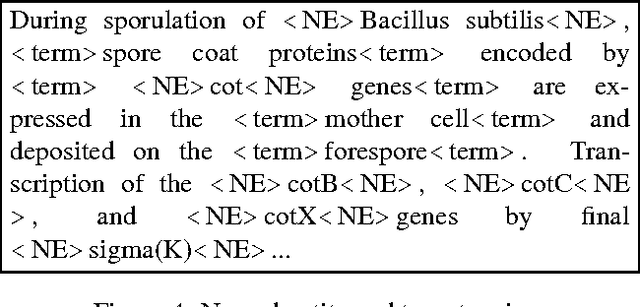

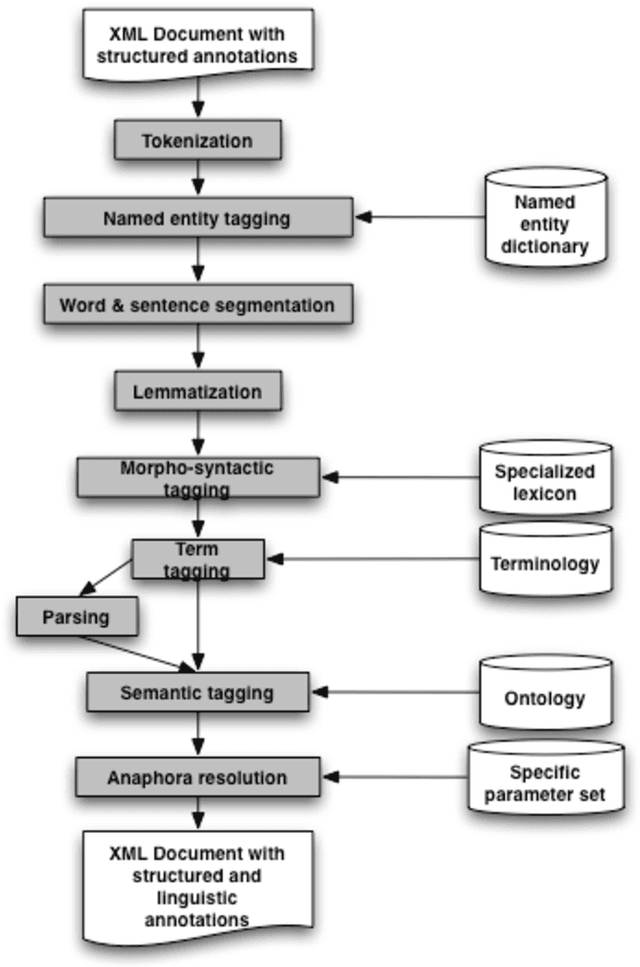
Abstract:The paper describes the ALVIS annotation format designed for the indexing of large collections of documents in topic-specific search engines. This paper is exemplified on the biological domain and on MedLine abstracts, as developing a specialized search engine for biologists is one of the ALVIS case studies. The ALVIS principle for linguistic annotations is based on existing works and standard propositions. We made the choice of stand-off annotations rather than inserted mark-up. Annotations are encoded as XML elements which form the linguistic subsection of the document record.
Event-based Information Extraction for the biomedical domain: the Caderige project
Sep 24, 2006
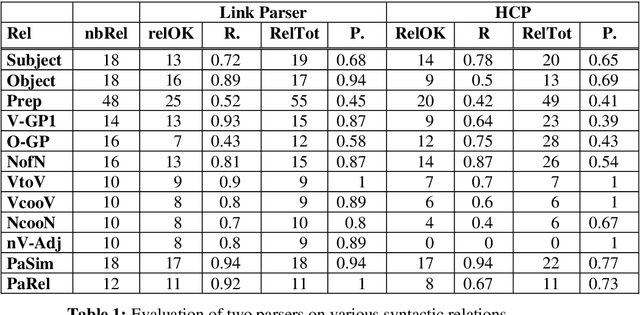
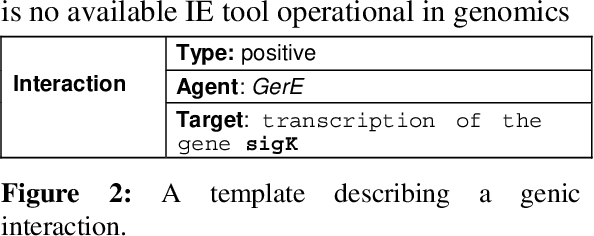
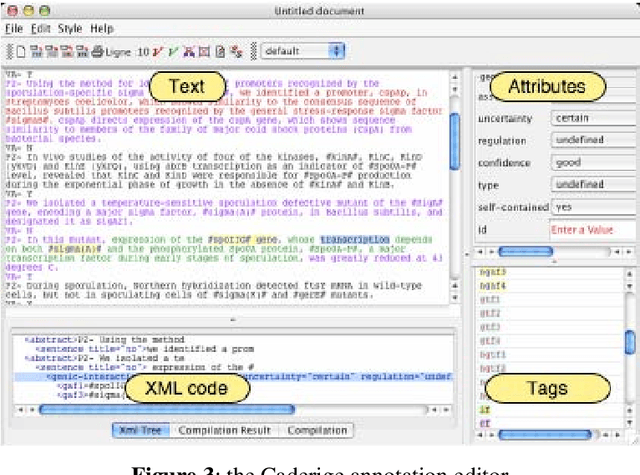
Abstract:This paper gives an overview of the Caderige project. This project involves teams from different areas (biology, machine learning, natural language processing) in order to develop high-level analysis tools for extracting structured information from biological bibliographical databases, especially Medline. The paper gives an overview of the approach and compares it to the state of the art.
Improving Term Extraction with Terminological Resources
Sep 06, 2006


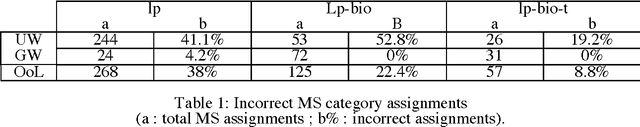
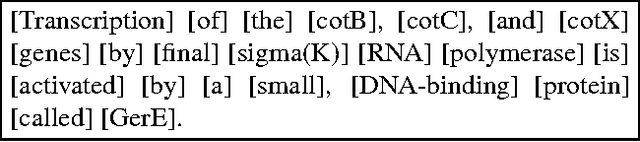
Abstract:Studies of different term extractors on a corpus of the biomedical domain revealed decreasing performances when applied to highly technical texts. The difficulty or impossibility of customising them to new domains is an additional limitation. In this paper, we propose to use external terminologies to influence generic linguistic data in order to augment the quality of the extraction. The tool we implemented exploits testified terms at different steps of the process: chunking, parsing and extraction of term candidates. Experiments reported here show that, using this method, more term candidates can be acquired with a higher level of reliability. We further describe the extraction process involving endogenous disambiguation implemented in the term extractor YaTeA.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge