Ciprian Catana
Martinos Center for Biomedical Imaging, Boston, Harvard Medical School, Boston
Direct Reconstruction of Linear Parametric Images from Dynamic PET Using Nonlocal Deep Image Prior
Jun 18, 2021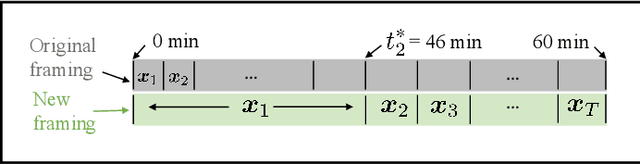
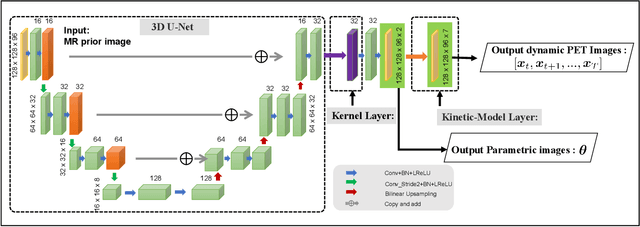
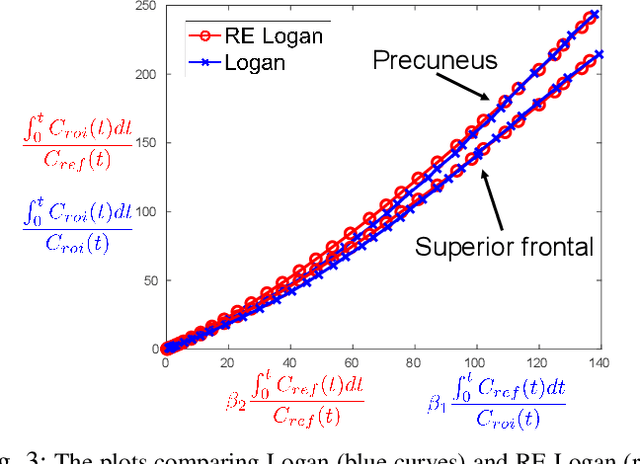
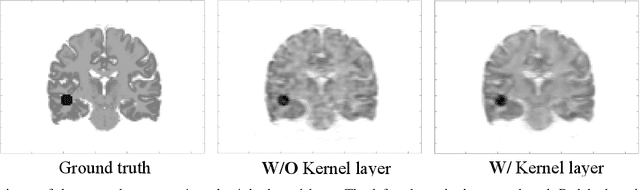
Abstract:Direct reconstruction methods have been developed to estimate parametric images directly from the measured PET sinograms by combining the PET imaging model and tracer kinetics in an integrated framework. Due to limited counts received, signal-to-noise-ratio (SNR) and resolution of parametric images produced by direct reconstruction frameworks are still limited. Recently supervised deep learning methods have been successfully applied to medical imaging denoising/reconstruction when large number of high-quality training labels are available. For static PET imaging, high-quality training labels can be acquired by extending the scanning time. However, this is not feasible for dynamic PET imaging, where the scanning time is already long enough. In this work, we proposed an unsupervised deep learning framework for direct parametric reconstruction from dynamic PET, which was tested on the Patlak model and the relative equilibrium Logan model. The patient's anatomical prior image, which is readily available from PET/CT or PET/MR scans, was supplied as the network input to provide a manifold constraint, and also utilized to construct a kernel layer to perform non-local feature denoising. The linear kinetic model was embedded in the network structure as a 1x1 convolution layer. The training objective function was based on the PET statistical model. Evaluations based on dynamic datasets of 18F-FDG and 11C-PiB tracers show that the proposed framework can outperform the traditional and the kernel method-based direct reconstruction methods.
Learning Personalized Representation for Inverse Problems in Medical Imaging Using Deep Neural Network
Jul 04, 2018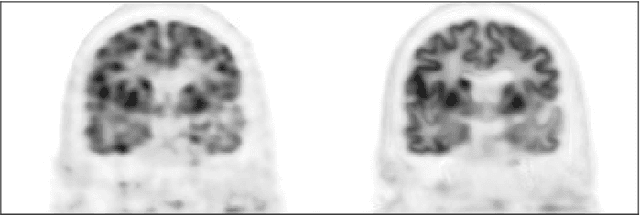

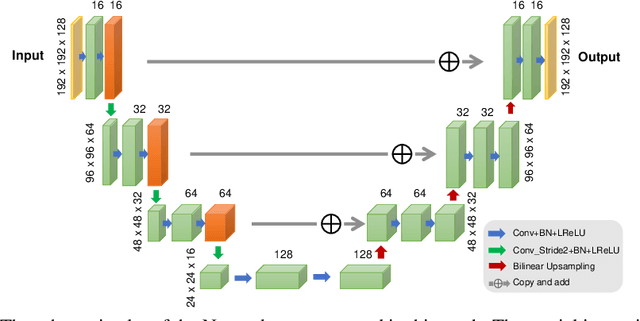
Abstract:Recently deep neural networks have been widely and successfully applied in computer vision tasks and attracted growing interests in medical imaging. One barrier for the application of deep neural networks to medical imaging is the need of large amounts of prior training pairs, which is not always feasible in clinical practice. In this work we propose a personalized representation learning framework where no prior training pairs are needed, but only the patient's own prior images. The representation is expressed using a deep neural network with the patient's prior images as network input. We then applied this novel image representation to inverse problems in medical imaging in which the original inverse problem was formulated as a constraint optimization problem and solved using the alternating direction method of multipliers (ADMM) algorithm. Anatomically guided brain positron emission tomography (PET) image reconstruction and image denoising were employed as examples to demonstrate the effectiveness of the proposed framework. Quantification results based on simulation and real datasets show that the proposed personalized representation framework outperform other widely adopted methods.
Kinetic Compressive Sensing
Mar 27, 2018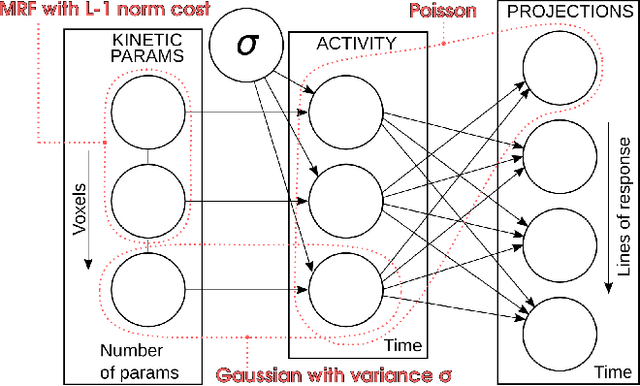
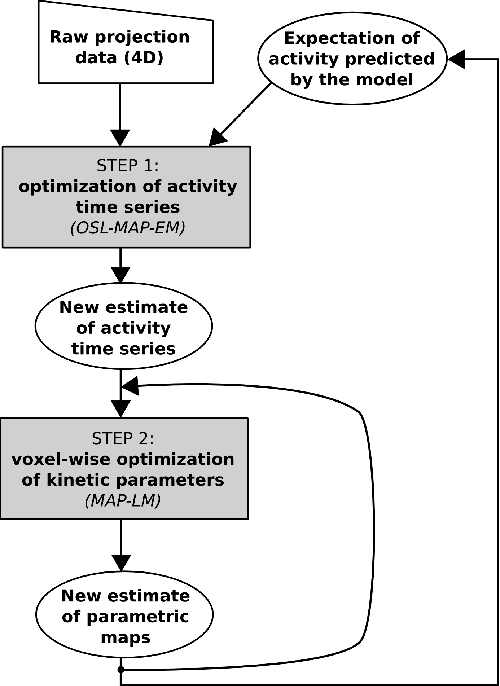

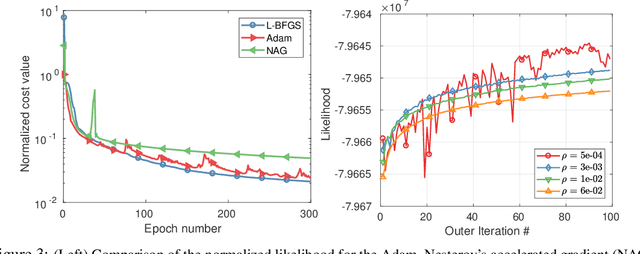
Abstract:Parametric images provide insight into the spatial distribution of physiological parameters, but they are often extremely noisy, due to low SNR of tomographic data. Direct estimation from projections allows accurate noise modeling, improving the results of post-reconstruction fitting. We propose a method, which we name kinetic compressive sensing (KCS), based on a hierarchical Bayesian model and on a novel reconstruction algorithm, that encodes sparsity of kinetic parameters. Parametric maps are reconstructed by maximizing the joint probability, with an Iterated Conditional Modes (ICM) approach, alternating the optimization of activity time series (OS-MAP-OSL), and kinetic parameters (MAP-LM). We evaluated the proposed algorithm on a simulated dynamic phantom: a bias/variance study confirmed how direct estimates can improve the quality of parametric maps over a post-reconstruction fitting, and showed how the novel sparsity prior can further reduce their variance, without affecting bias. Real FDG PET human brain data (Siemens mMR, 40min) images were also processed. Results enforced how the proposed KCS-regularized direct method can produce spatially coherent images and parametric maps, with lower spatial noise and better tissue contrast. A GPU-based open source implementation of the algorithm is provided.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge