Stefano Cerri
A large-scale heterogeneous 3D magnetic resonance brain imaging dataset for self-supervised learning
Jun 17, 2025Abstract:We present FOMO60K, a large-scale, heterogeneous dataset of 60,529 brain Magnetic Resonance Imaging (MRI) scans from 13,900 sessions and 11,187 subjects, aggregated from 16 publicly available sources. The dataset includes both clinical- and research-grade images, multiple MRI sequences, and a wide range of anatomical and pathological variability, including scans with large brain anomalies. Minimal preprocessing was applied to preserve the original image characteristics while reducing barriers to entry for new users. Accompanying code for self-supervised pretraining and finetuning is provided. FOMO60K is intended to support the development and benchmarking of self-supervised learning methods in medical imaging at scale.
Quantifying white matter hyperintensity and brain volumes in heterogeneous clinical and low-field portable MRI
Dec 08, 2023
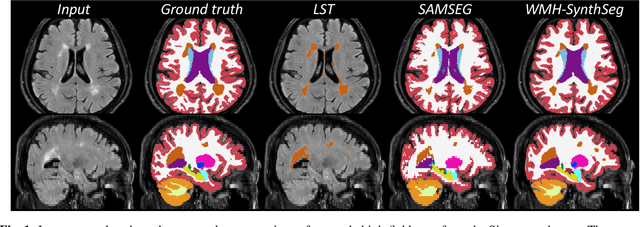
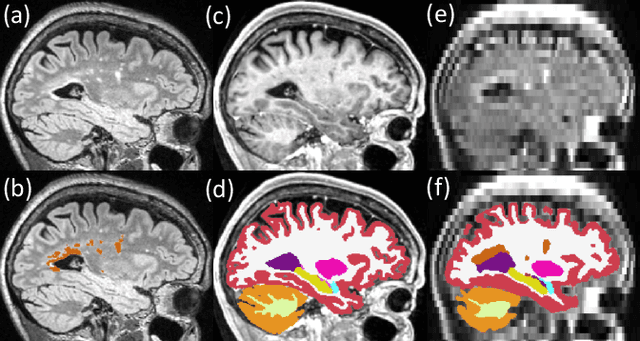

Abstract:Brain atrophy and white matter hyperintensity (WMH) are critical neuroimaging features for ascertaining brain injury in cerebrovascular disease and multiple sclerosis. Automated segmentation and quantification is desirable but existing methods require high-resolution MRI with good signal-to-noise ratio (SNR). This precludes application to clinical and low-field portable MRI (pMRI) scans, thus hampering large-scale tracking of atrophy and WMH progression, especially in underserved areas where pMRI has huge potential. Here we present a method that segments white matter hyperintensity and 36 brain regions from scans of any resolution and contrast (including pMRI) without retraining. We show results on six public datasets and on a private dataset with paired high- and low-field scans (3T and 64mT), where we attain strong correlation between the WMH ($\rho$=.85) and hippocampal volumes (r=.89) estimated at both fields. Our method is publicly available as part of FreeSurfer, at: http://surfer.nmr.mgh.harvard.edu/fswiki/WMH-SynthSeg.
A Lightweight Causal Model for Interpretable Subject-level Prediction
Jun 19, 2023



Abstract:Recent years have seen a growing interest in methods for predicting a variable of interest, such as a subject's diagnosis, from medical images. Methods based on discriminative modeling excel at making accurate predictions, but are challenged in their ability to explain their decisions in anatomically meaningful terms. In this paper, we propose a simple technique for single-subject prediction that is inherently interpretable. It augments the generative models used in classical human brain mapping techniques, in which cause-effect relations can be encoded, with a multivariate noise model that captures dominant spatial correlations. Experiments demonstrate that the resulting model can be efficiently inverted to make accurate subject-level predictions, while at the same time offering intuitive causal explanations of its inner workings. The method is easy to use: training is fast for typical training set sizes, and only a single hyperparameter needs to be set by the user. Our code is available at https://github.com/chiara-mauri/Interpretable-subject-level-prediction.
An Open-Source Tool for Longitudinal Whole-Brain and White Matter Lesion Segmentation
Jul 10, 2022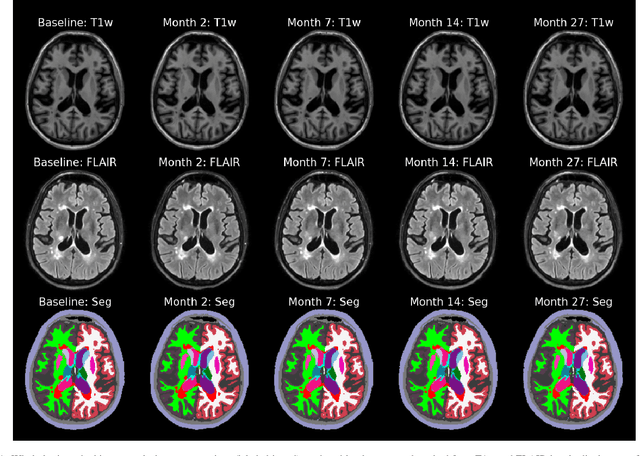
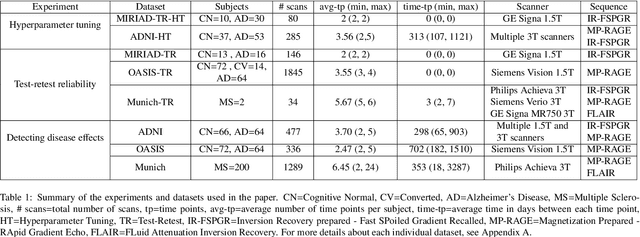
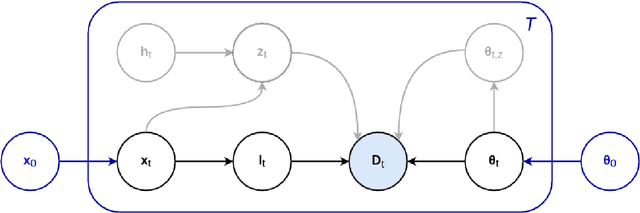
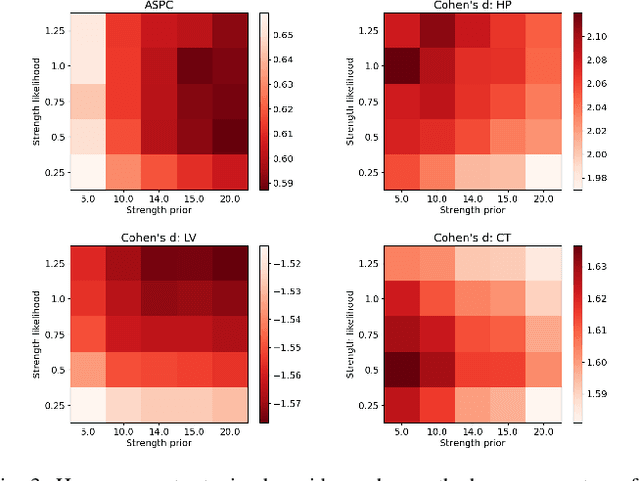
Abstract:In this paper we describe and validate a longitudinal method for whole-brain segmentation of longitudinal MRI scans. It builds upon an existing whole-brain segmentation method that can handle multi-contrast data and robustly analyze images with white matter lesions. This method is here extended with subject-specific latent variables that encourage temporal consistency between its segmentation results, enabling it to better track subtle morphological changes in dozens of neuroanatomical structures and white matter lesions. We validate the proposed method on multiple datasets of control subjects and patients suffering from Alzheimer's disease and multiple sclerosis, and compare its results against those obtained with its original cross-sectional formulation and two benchmark longitudinal methods. The results indicate that the method attains a higher test-retest reliability, while being more sensitive to longitudinal disease effect differences between patient groups. An implementation is publicly available as part of the open-source neuroimaging package FreeSurfer.
Prediction of MGMT Methylation Status of Glioblastoma using Radiomics and Latent Space Shape Features
Sep 25, 2021


Abstract:In this paper we propose a method for predicting the status of MGMT promoter methylation in high-grade gliomas. From the available MR images, we segment the tumor using deep convolutional neural networks and extract both radiomic features and shape features learned by a variational autoencoder. We implemented a standard machine learning workflow to obtain predictions, consisting of feature selection followed by training of a random forest classification model. We trained and evaluated our method on the RSNA-ASNR-MICCAI BraTS 2021 challenge dataset and submitted our predictions to the challenge.
Predicting survival of glioblastoma from automatic whole-brain and tumor segmentation of MR images
Sep 25, 2021



Abstract:Survival prediction models can potentially be used to guide treatment of glioblastoma patients. However, currently available MR imaging biomarkers holding prognostic information are often challenging to interpret, have difficulties generalizing across data acquisitions, or are only applicable to pre-operative MR data. In this paper we aim to address these issues by introducing novel imaging features that can be automatically computed from MR images and fed into machine learning models to predict patient survival. The features we propose have a direct biological interpretation: They measure the deformation caused by the tumor on the surrounding brain structures, comparing the shape of various structures in the patient's brain to their expected shape in healthy individuals. To obtain the required segmentations, we use an automatic method that is contrast-adaptive and robust to missing modalities, making the features generalizable across scanners and imaging protocols. Since the features we propose do not depend on characteristics of the tumor region itself, they are also applicable to post-operative images, which have been much less studied in the context of survival prediction. Using experiments involving both pre- and post-operative data, we show that the proposed features carry prognostic value in terms of overall- and progression-free survival, over and above that of conventional non-imaging features.
A Longitudinal Method for Simultaneous Whole-Brain and Lesion Segmentation in Multiple Sclerosis
Sep 15, 2020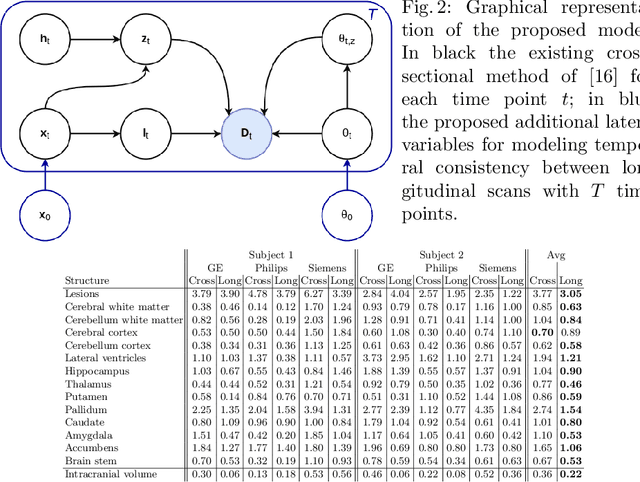
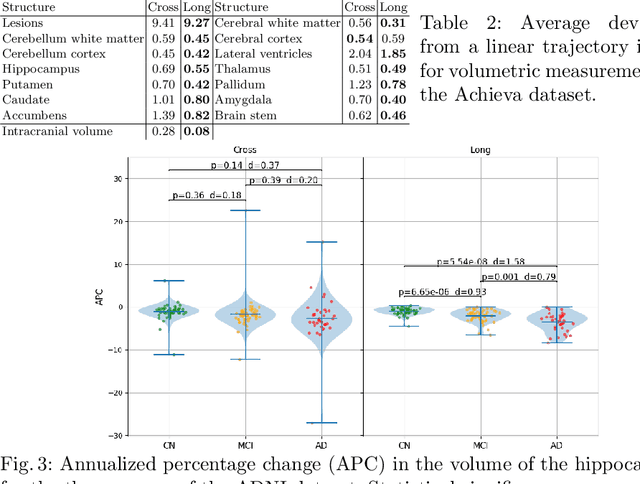
Abstract:In this paper we propose a novel method for the segmentation of longitudinal brain MRI scans of patients suffering from Multiple Sclerosis. The method builds upon an existing cross-sectional method for simultaneous whole-brain and lesion segmentation, introducing subject-specific latent variables to encourage temporal consistency between longitudinal scans. It is very generally applicable, as it does not make any prior assumptions on the scanner, the MRI protocol, or the number and timing of longitudinal follow-up scans. Preliminary experiments on three longitudinal datasets indicate that the proposed method produces more reliable segmentations and detects disease effects better than the cross-sectional method it is based upon.
A Contrast-Adaptive Method for Simultaneous Whole-Brain and Lesion Segmentation in Multiple Sclerosis
May 11, 2020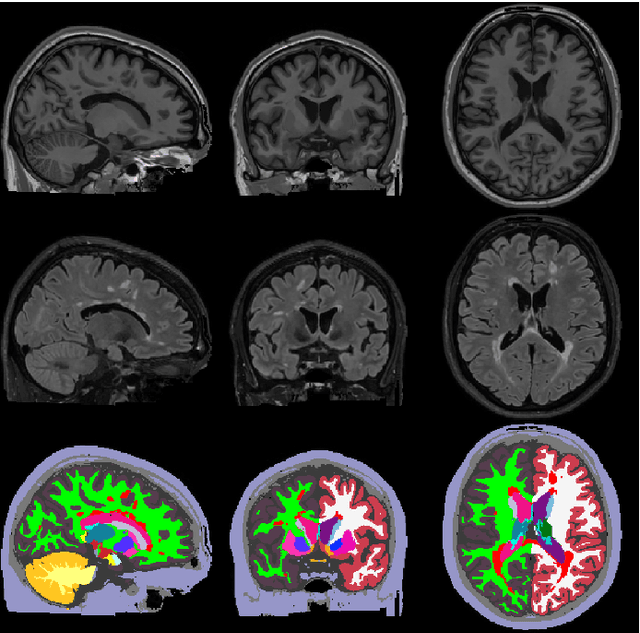
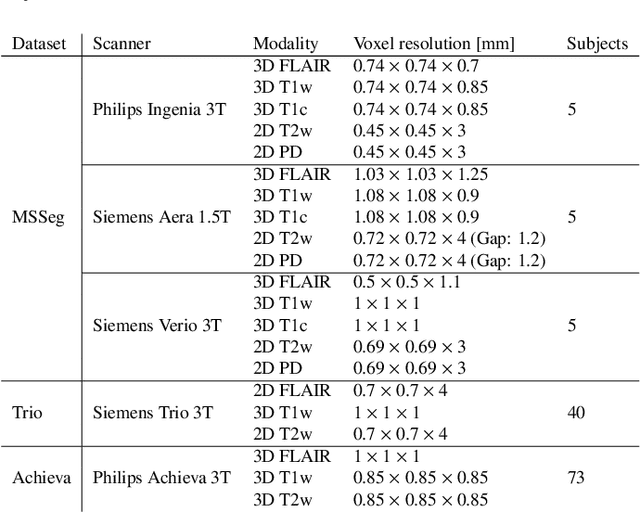
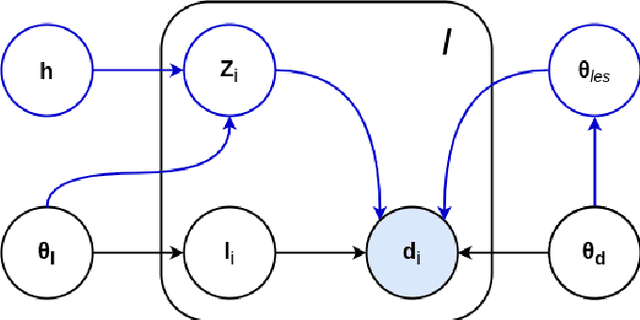
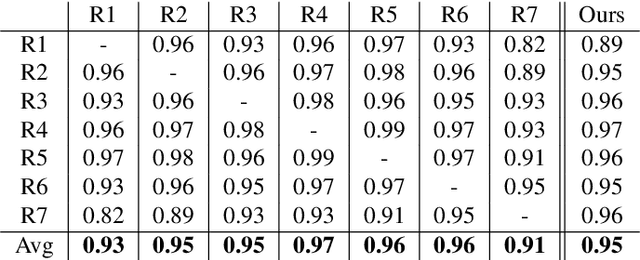
Abstract:Here we present a method for the simultaneous segmentation of white matter lesions and normal-appearing neuroanatomical structures from multi-contrast brain MRI scans of multiple sclerosis patients. The method integrates a novel model for white matter lesions into a previously validated generative model for whole-brain segmentation. By using separate models for the shape of anatomical structures and their appearance in MRI, the algorithm can adapt to data acquired with different scanners and imaging protocols without retraining. We validate the method using three disparate datasets, showing state-of-the-art performance in white matter lesion segmentation while simultaneously segmenting dozens of other brain structures. We further demonstrate that the contrast-adaptive method can also be applied robustly to MRI scans of healthy controls, and replicate previously documented atrophy patterns in deep gray matter structures in MS. The algorithm is publicly available as part of the open-source neuroimaging package FreeSurfer.
Semi-Supervised Variational Autoencoder for Survival Prediction
Oct 10, 2019



Abstract:In this paper we propose a semi-supervised variational autoencoder for classification of overall survival groups from tumor segmentation masks. The model can use the output of any tumor segmentation algorithm, removing all assumptions on the scanning platform and the specific type of pulse sequences used, thereby increasing its generalization properties. Due to its semi-supervised nature, the method can learn to classify survival time by using a relatively small number of labeled subjects. We validate our model on the publicly available dataset from the Multimodal Brain Tumor Segmentation Challenge (BraTS) 2019.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge