Ian Law
Department of Clinical Physiology, Nuclear Medicine and PET, Copenhagen University Hospital Rigshospitalet, Denmark
Predicting survival of glioblastoma from automatic whole-brain and tumor segmentation of MR images
Sep 25, 2021



Abstract:Survival prediction models can potentially be used to guide treatment of glioblastoma patients. However, currently available MR imaging biomarkers holding prognostic information are often challenging to interpret, have difficulties generalizing across data acquisitions, or are only applicable to pre-operative MR data. In this paper we aim to address these issues by introducing novel imaging features that can be automatically computed from MR images and fed into machine learning models to predict patient survival. The features we propose have a direct biological interpretation: They measure the deformation caused by the tumor on the surrounding brain structures, comparing the shape of various structures in the patient's brain to their expected shape in healthy individuals. To obtain the required segmentations, we use an automatic method that is contrast-adaptive and robust to missing modalities, making the features generalizable across scanners and imaging protocols. Since the features we propose do not depend on characteristics of the tumor region itself, they are also applicable to post-operative images, which have been much less studied in the context of survival prediction. Using experiments involving both pre- and post-operative data, we show that the proposed features carry prognostic value in terms of overall- and progression-free survival, over and above that of conventional non-imaging features.
A Modality-Adaptive Method for Segmenting Brain Tumors and Organs-at-Risk in Radiation Therapy Planning
Aug 15, 2018
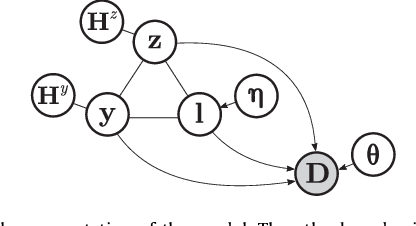
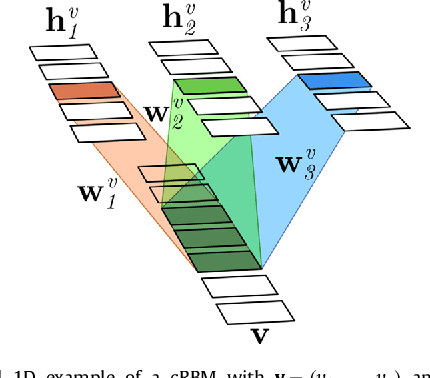

Abstract:In this paper we present a method for simultaneously segmenting brain tumors and an extensive set of organs-at-risk for radiation therapy planning of glioblastomas. The method combines a contrast-adaptive generative model for whole-brain segmentation with a new spatial regularization model of tumor shape using convolutional restricted Boltzmann machines. We demonstrate experimentally that the method is able to adapt to image acquisitions that differ substantially from any available training data, ensuring its applicability across treatment sites; that its tumor segmentation accuracy is comparable to that of the current state of the art; and that it captures most organs-at-risk sufficiently well for radiation therapy planning purposes. The proposed method may be a valuable step towards automating the delineation of brain tumors and organs-at-risk in glioblastoma patients undergoing radiation therapy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge