Dominik S. Meier
Using novel data and ensemble models to improve automated labeling of Sustainable Development Goals
Feb 01, 2023Abstract:A number of labeling systems based on text have been proposed to help monitor work on the United Nations (UN) Sustainable Development Goals (SDGs). Here, we present a systematic comparison of systems using a variety of text sources and show that systems differ considerably in their specificity (i.e., true-positive rate) and sensitivity (i.e., true-negative rate), have systematic biases (e.g., are more sensitive to specific SDGs relative to others), and are susceptible to the type and amount of text analyzed. We then show that an ensemble model that pools labeling systems alleviates some of these limitations, exceeding the labeling performance of all currently available systems. We conclude that researchers and policymakers should care about the choice of labeling system and that ensemble methods should be favored when drawing conclusions about the absolute and relative prevalence of work on the SDGs based on automated methods.
text2sdg: An open-source solution to monitoring sustainable development goals from text
Oct 20, 2021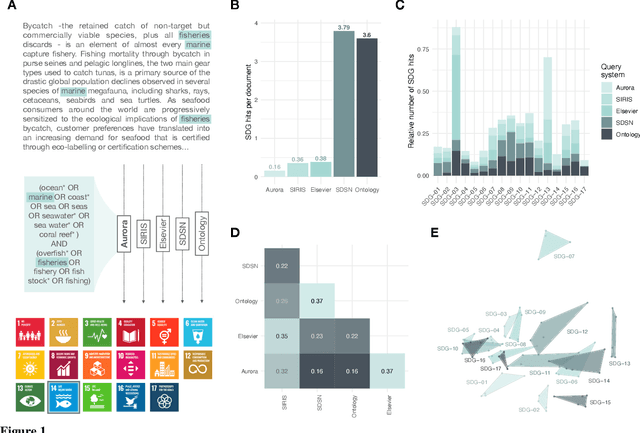
Abstract:Monitoring progress on the United Nations Sustainable Development Goals (SDGs) is important for both academic and non-academic organizations. Existing approaches to monitoring SDGs have focused on specific data types, namely, publications listed in proprietary research databases. We present the text2sdg R package, a user-friendly, open-source package that detects SDGs in any kind of text data using several different query systems from any text source. The text2sdg package thereby facilitates the monitoring of SDGs for a wide array of text sources and provides a much-needed basis for validating and improving extant methods to detect SDGs from text.
A Contrast-Adaptive Method for Simultaneous Whole-Brain and Lesion Segmentation in Multiple Sclerosis
May 11, 2020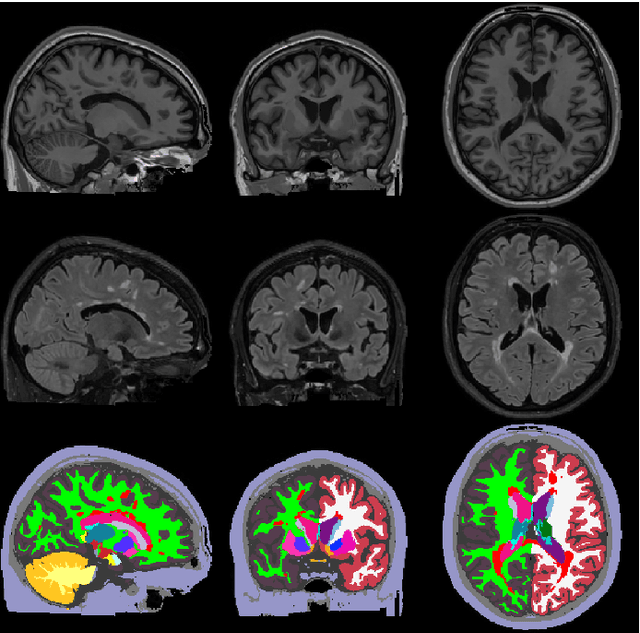
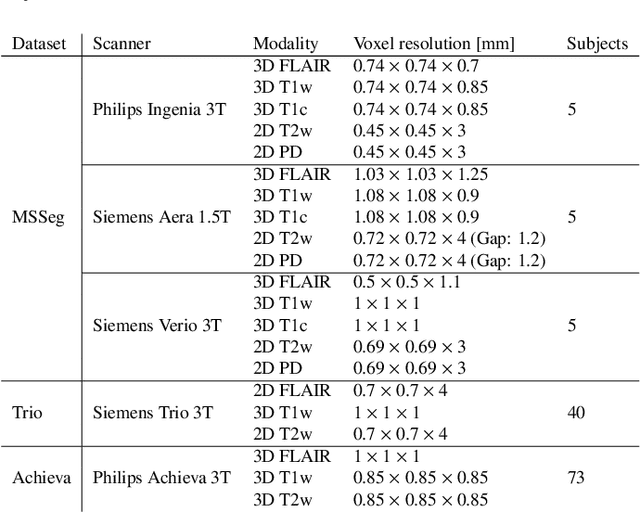
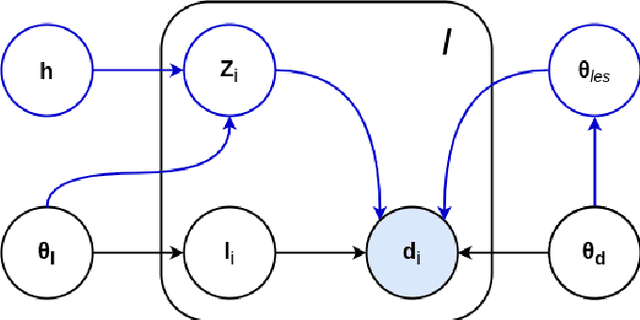
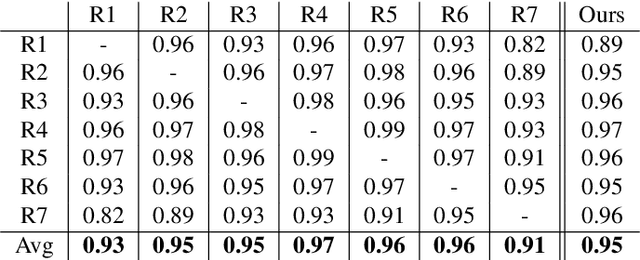
Abstract:Here we present a method for the simultaneous segmentation of white matter lesions and normal-appearing neuroanatomical structures from multi-contrast brain MRI scans of multiple sclerosis patients. The method integrates a novel model for white matter lesions into a previously validated generative model for whole-brain segmentation. By using separate models for the shape of anatomical structures and their appearance in MRI, the algorithm can adapt to data acquired with different scanners and imaging protocols without retraining. We validate the method using three disparate datasets, showing state-of-the-art performance in white matter lesion segmentation while simultaneously segmenting dozens of other brain structures. We further demonstrate that the contrast-adaptive method can also be applied robustly to MRI scans of healthy controls, and replicate previously documented atrophy patterns in deep gray matter structures in MS. The algorithm is publicly available as part of the open-source neuroimaging package FreeSurfer.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge