Marianne Rakic
Pancakes: Consistent Multi-Protocol Image Segmentation Across Biomedical Domains
Dec 15, 2025Abstract:A single biomedical image can be meaningfully segmented in multiple ways, depending on the desired application. For instance, a brain MRI can be segmented according to tissue types, vascular territories, broad anatomical regions, fine-grained anatomy, or pathology, etc. Existing automatic segmentation models typically either (1) support only a single protocol, the one they were trained on, or (2) require labor-intensive manual prompting to specify the desired segmentation. We introduce Pancakes, a framework that, given a new image from a previously unseen domain, automatically generates multi-label segmentation maps for multiple plausible protocols, while maintaining semantic consistency across related images. Pancakes introduces a new problem formulation that is not currently attainable by existing foundation models. In a series of experiments on seven held-out datasets, we demonstrate that our model can significantly outperform existing foundation models in producing several plausible whole-image segmentations, that are semantically coherent across images.
AtlasMorph: Learning conditional deformable templates for brain MRI
Nov 17, 2025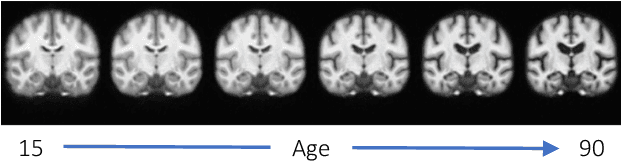
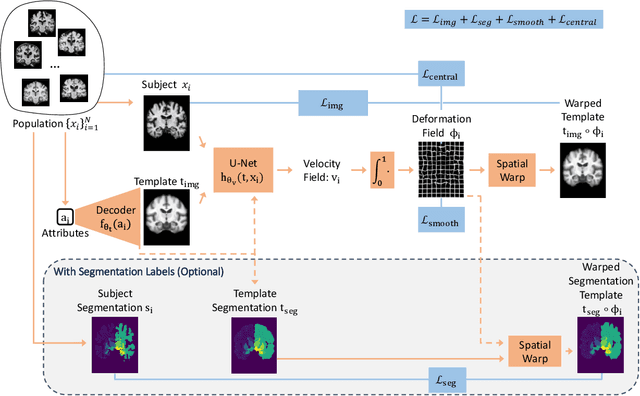
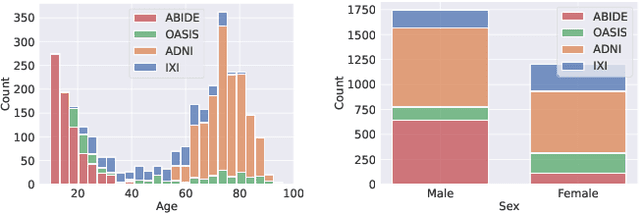
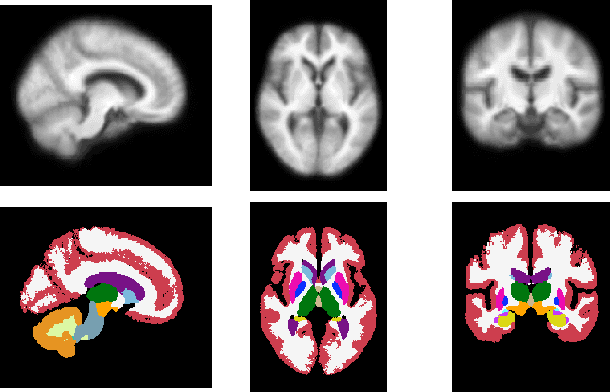
Abstract:Deformable templates, or atlases, are images that represent a prototypical anatomy for a population, and are often enhanced with probabilistic anatomical label maps. They are commonly used in medical image analysis for population studies and computational anatomy tasks such as registration and segmentation. Because developing a template is a computationally expensive process, relatively few templates are available. As a result, analysis is often conducted with sub-optimal templates that are not truly representative of the study population, especially when there are large variations within this population. We propose a machine learning framework that uses convolutional registration neural networks to efficiently learn a function that outputs templates conditioned on subject-specific attributes, such as age and sex. We also leverage segmentations, when available, to produce anatomical segmentation maps for the resulting templates. The learned network can also be used to register subject images to the templates. We demonstrate our method on a compilation of 3D brain MRI datasets, and show that it can learn high-quality templates that are representative of populations. We find that annotated conditional templates enable better registration than their unlabeled unconditional counterparts, and outperform other templates construction methods.
MultiMorph: On-demand Atlas Construction
Mar 31, 2025Abstract:We present MultiMorph, a fast and efficient method for constructing anatomical atlases on the fly. Atlases capture the canonical structure of a collection of images and are essential for quantifying anatomical variability across populations. However, current atlas construction methods often require days to weeks of computation, thereby discouraging rapid experimentation. As a result, many scientific studies rely on suboptimal, precomputed atlases from mismatched populations, negatively impacting downstream analyses. MultiMorph addresses these challenges with a feedforward model that rapidly produces high-quality, population-specific atlases in a single forward pass for any 3D brain dataset, without any fine-tuning or optimization. MultiMorph is based on a linear group-interaction layer that aggregates and shares features within the group of input images. Further, by leveraging auxiliary synthetic data, MultiMorph generalizes to new imaging modalities and population groups at test-time. Experimentally, MultiMorph outperforms state-of-the-art optimization-based and learning-based atlas construction methods in both small and large population settings, with a 100-fold reduction in time. This makes MultiMorph an accessible framework for biomedical researchers without machine learning expertise, enabling rapid, high-quality atlas generation for diverse studies.
Tyche: Stochastic In-Context Learning for Medical Image Segmentation
Jan 24, 2024



Abstract:Existing learning-based solutions to medical image segmentation have two important shortcomings. First, for most new segmentation task, a new model has to be trained or fine-tuned. This requires extensive resources and machine learning expertise, and is therefore often infeasible for medical researchers and clinicians. Second, most existing segmentation methods produce a single deterministic segmentation mask for a given image. In practice however, there is often considerable uncertainty about what constitutes the correct segmentation, and different expert annotators will often segment the same image differently. We tackle both of these problems with Tyche, a model that uses a context set to generate stochastic predictions for previously unseen tasks without the need to retrain. Tyche differs from other in-context segmentation methods in two important ways. (1) We introduce a novel convolution block architecture that enables interactions among predictions. (2) We introduce in-context test-time augmentation, a new mechanism to provide prediction stochasticity. When combined with appropriate model design and loss functions, Tyche can predict a set of plausible diverse segmentation candidates for new or unseen medical images and segmentation tasks without the need to retrain.
ScribblePrompt: Fast and Flexible Interactive Segmentation for Any Medical Image
Dec 12, 2023Abstract:Semantic medical image segmentation is a crucial part of both scientific research and clinical care. With enough labelled data, deep learning models can be trained to accurately automate specific medical image segmentation tasks. However, manually segmenting images to create training data is highly labor intensive. In this paper, we present ScribblePrompt, an interactive segmentation framework for medical imaging that enables human annotators to segment unseen structures using scribbles, clicks, and bounding boxes. Scribbles are an intuitive and effective form of user interaction for complex tasks, however most existing methods focus on click-based interactions. We introduce algorithms for simulating realistic scribbles that enable training models that are amenable to multiple types of interaction. To achieve generalization to new tasks, we train on a diverse collection of 65 open-access biomedical datasets -- using both real and synthetic labels. We test ScribblePrompt on multiple network architectures and unseen datasets, and demonstrate that it can be used in real-time on a single CPU. We evaluate ScribblePrompt using manually-collected scribbles, simulated interactions, and a user study. ScribblePrompt outperforms existing methods in all our evaluations. In the user study, ScribblePrompt reduced annotation time by 28% while improving Dice by 15% compared to existing methods. We showcase ScribblePrompt in an online demo and provide code at https://scribbleprompt.csail.mit.edu
Anatomical Predictions using Subject-Specific Medical Data
May 29, 2020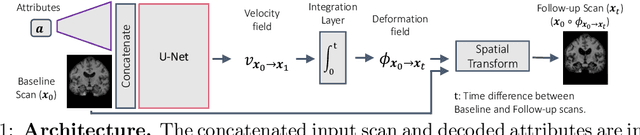
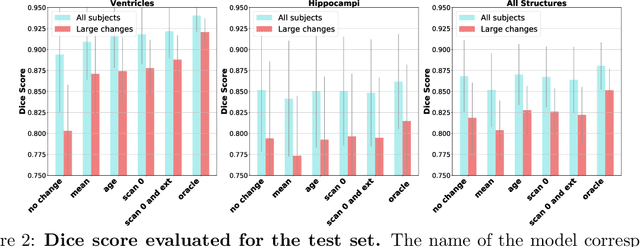
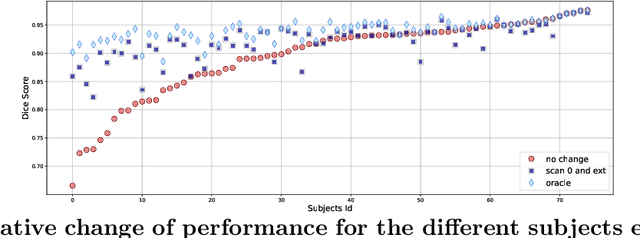
Abstract:Changes over time in brain anatomy can provide important insight for treatment design or scientific analyses. We present a method that predicts how a brain MRI for an individual will change over time. We model changes using a diffeomorphic deformation field that we predict using function using convolutional neural networks. Given a predicted deformation field, a baseline scan can be warped to give a prediction of the brain scan at a future time. We demonstrate the method using the ADNI cohort, and analyze how performance is affected by model variants and the subject-specific information provided. We show that the model provides good predictions and that external clinical data can improve predictions.
Learning Conditional Deformable Templates with Convolutional Networks
Aug 07, 2019
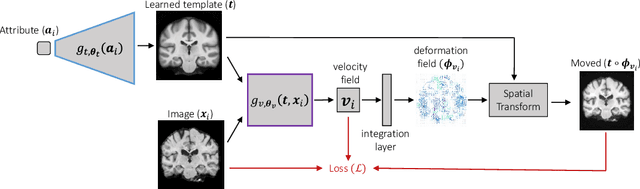
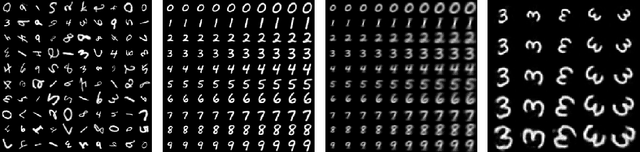

Abstract:We develop a learning framework for building deformable templates, which play a fundamental role in many image analysis and computational anatomy tasks. Conventional methods for template creation and image alignment to the template have undergone decades of rich technical development. In these frameworks, templates are constructed using an iterative process of template estimation and alignment, which is often computationally very expensive. Due in part to this shortcoming, most methods compute a single template for the entire population of images, or a few templates for specific sub-groups of the data. In this work, we present a probabilistic model and efficient learning strategy that yields either universal or conditional templates, jointly with a neural network that provides efficient alignment of the images to these templates. We demonstrate the usefulness of this method on a variety of domains, with a special focus on neuroimaging. This is particularly useful for clinical applications where a pre-existing template does not exist, or creating a new one with traditional methods can be prohibitively expensive. Our code and atlases are available online as part of the VoxelMorph library at http://voxelmorph.csail.mit.edu.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge