Jose Javier Gonzalez Ortiz
MultiverSeg: Scalable Interactive Segmentation of Biomedical Imaging Datasets with In-Context Guidance
Dec 19, 2024Abstract:Medical researchers and clinicians often need to perform novel segmentation tasks on a set of related images. Existing methods for segmenting a new dataset are either interactive, requiring substantial human effort for each image, or require an existing set of manually labeled images. We introduce a system, MultiverSeg, that enables practitioners to rapidly segment an entire new dataset without requiring access to any existing labeled data from that task or domain. Along with the image to segment, the model takes user interactions such as clicks, bounding boxes or scribbles as input, and predicts a segmentation. As the user segments more images, those images and segmentations become additional inputs to the model, providing context. As the context set of labeled images grows, the number of interactions required to segment each new image decreases. We demonstrate that MultiverSeg enables users to interactively segment new datasets efficiently, by amortizing the number of interactions per image to achieve an accurate segmentation. Compared to using a state-of-the-art interactive segmentation method, using MultiverSeg reduced the total number of scribble steps by 53% and clicks by 36% to achieve 90% Dice on sets of images from unseen tasks. We release code and model weights at https://multiverseg.csail.mit.edu
Tyche: Stochastic In-Context Learning for Medical Image Segmentation
Jan 24, 2024



Abstract:Existing learning-based solutions to medical image segmentation have two important shortcomings. First, for most new segmentation task, a new model has to be trained or fine-tuned. This requires extensive resources and machine learning expertise, and is therefore often infeasible for medical researchers and clinicians. Second, most existing segmentation methods produce a single deterministic segmentation mask for a given image. In practice however, there is often considerable uncertainty about what constitutes the correct segmentation, and different expert annotators will often segment the same image differently. We tackle both of these problems with Tyche, a model that uses a context set to generate stochastic predictions for previously unseen tasks without the need to retrain. Tyche differs from other in-context segmentation methods in two important ways. (1) We introduce a novel convolution block architecture that enables interactions among predictions. (2) We introduce in-context test-time augmentation, a new mechanism to provide prediction stochasticity. When combined with appropriate model design and loss functions, Tyche can predict a set of plausible diverse segmentation candidates for new or unseen medical images and segmentation tasks without the need to retrain.
Non-Proportional Parametrizations for Stable Hypernetwork Learning
Apr 15, 2023



Abstract:Hypernetworks are neural networks that generate the parameters of another neural network. In many scenarios, current hypernetwork training strategies are unstable, and convergence is often far slower than for non-hypernetwork models. We show that this problem is linked to an issue that arises when using common choices of hypernetwork architecture and initialization. We demonstrate analytically and experimentally how this numerical issue can lead to an instability during training that slows, and sometimes even prevents, convergence. We also demonstrate that popular deep learning normalization strategies fail to address these issues. We then propose a solution to the problem based on a revised hypernetwork formulation that uses non-proportional additive parametrizations. We test the proposed reparametrization on several tasks, and demonstrate that it consistently leads to more stable training, achieving faster convergence.
UniverSeg: Universal Medical Image Segmentation
Apr 12, 2023



Abstract:While deep learning models have become the predominant method for medical image segmentation, they are typically not capable of generalizing to unseen segmentation tasks involving new anatomies, image modalities, or labels. Given a new segmentation task, researchers generally have to train or fine-tune models, which is time-consuming and poses a substantial barrier for clinical researchers, who often lack the resources and expertise to train neural networks. We present UniverSeg, a method for solving unseen medical segmentation tasks without additional training. Given a query image and example set of image-label pairs that define a new segmentation task, UniverSeg employs a new Cross-Block mechanism to produce accurate segmentation maps without the need for additional training. To achieve generalization to new tasks, we have gathered and standardized a collection of 53 open-access medical segmentation datasets with over 22,000 scans, which we refer to as MegaMedical. We used this collection to train UniverSeg on a diverse set of anatomies and imaging modalities. We demonstrate that UniverSeg substantially outperforms several related methods on unseen tasks, and thoroughly analyze and draw insights about important aspects of the proposed system. The UniverSeg source code and model weights are freely available at https://universeg.csail.mit.edu
Amortized Learning of Dynamic Feature Scaling for Image Segmentation
Apr 11, 2023Abstract:Convolutional neural networks (CNN) have become the predominant model for image segmentation tasks. Most CNN segmentation architectures resize spatial dimensions by a fixed factor of two to aggregate spatial context. Recent work has explored using other resizing factors to improve model accuracy for specific applications. However, finding the appropriate rescaling factor most often involves training a separate network for many different factors and comparing the performance of each model. The computational burden of these models means that in practice it is rarely done, and when done only a few different scaling factors are considered. In this work, we present a hypernetwork strategy that can be used to easily and rapidly generate the Pareto frontier for the trade-off between accuracy and efficiency as the rescaling factor varies. We show how to train a single hypernetwork that generates CNN parameters conditioned on a rescaling factor. This enables a user to quickly choose a rescaling factor that appropriately balances accuracy and computational efficiency for their particular needs. We focus on image segmentation tasks, and demonstrate the value of this approach across various domains. We also find that, for a given rescaling factor, our single hypernetwork outperforms CNNs trained with fixed rescaling factors.
Trade-offs of Local SGD at Scale: An Empirical Study
Oct 15, 2021
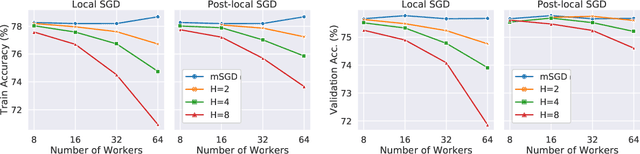
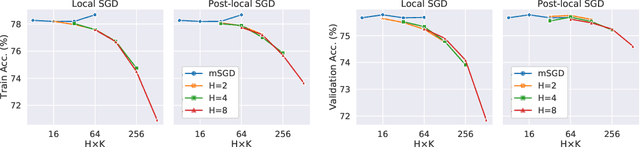
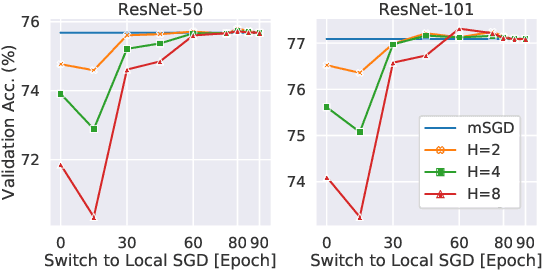
Abstract:As datasets and models become increasingly large, distributed training has become a necessary component to allow deep neural networks to train in reasonable amounts of time. However, distributed training can have substantial communication overhead that hinders its scalability. One strategy for reducing this overhead is to perform multiple unsynchronized SGD steps independently on each worker between synchronization steps, a technique known as local SGD. We conduct a comprehensive empirical study of local SGD and related methods on a large-scale image classification task. We find that performing local SGD comes at a price: lower communication costs (and thereby faster training) are accompanied by lower accuracy. This finding is in contrast from the smaller-scale experiments in prior work, suggesting that local SGD encounters challenges at scale. We further show that incorporating the slow momentum framework of Wang et al. (2020) consistently improves accuracy without requiring additional communication, hinting at future directions for potentially escaping this trade-off.
What is the State of Neural Network Pruning?
Mar 06, 2020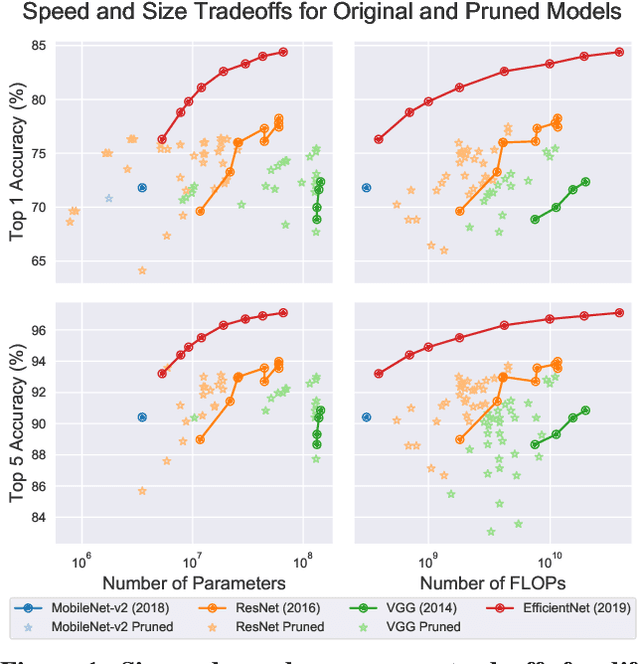
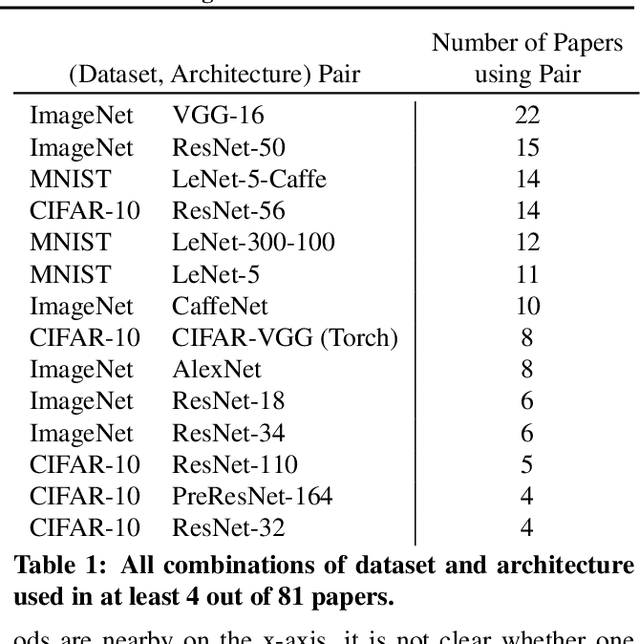
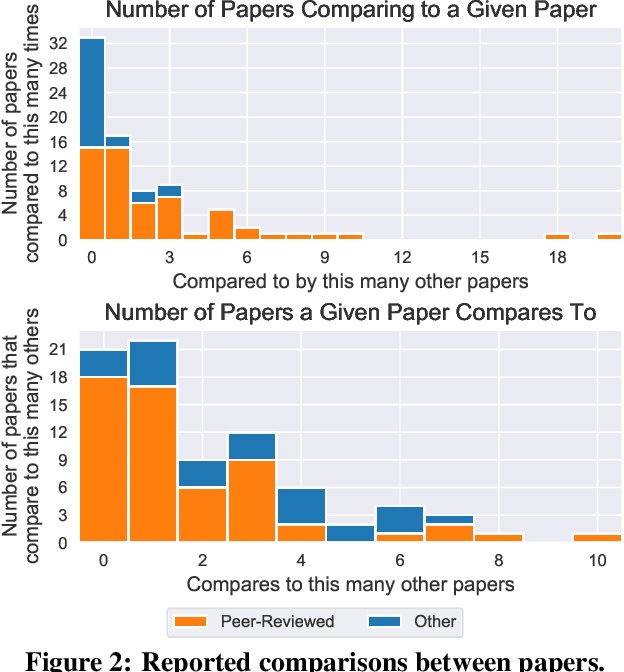
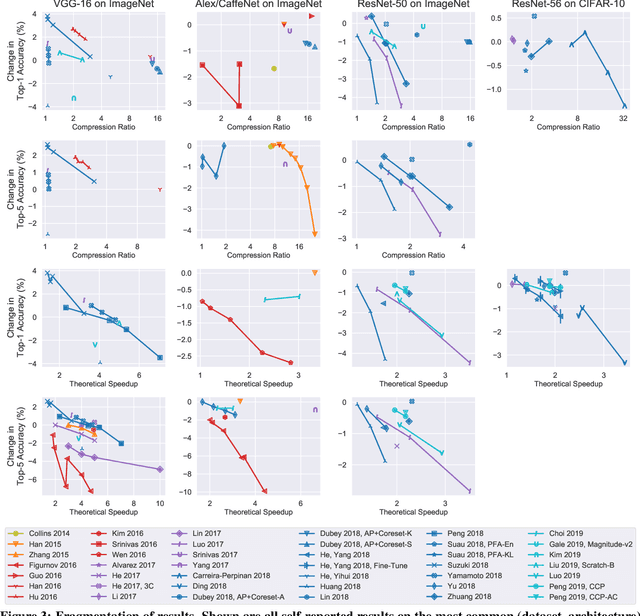
Abstract:Neural network pruning---the task of reducing the size of a network by removing parameters---has been the subject of a great deal of work in recent years. We provide a meta-analysis of the literature, including an overview of approaches to pruning and consistent findings in the literature. After aggregating results across 81 papers and pruning hundreds of models in controlled conditions, our clearest finding is that the community suffers from a lack of standardized benchmarks and metrics. This deficiency is substantial enough that it is hard to compare pruning techniques to one another or determine how much progress the field has made over the past three decades. To address this situation, we identify issues with current practices, suggest concrete remedies, and introduce ShrinkBench, an open-source framework to facilitate standardized evaluations of pruning methods. We use ShrinkBench to compare various pruning techniques and show that its comprehensive evaluation can prevent common pitfalls when comparing pruning methods.
Image segmentation of liver stage malaria infection with spatial uncertainty sampling
Nov 30, 2019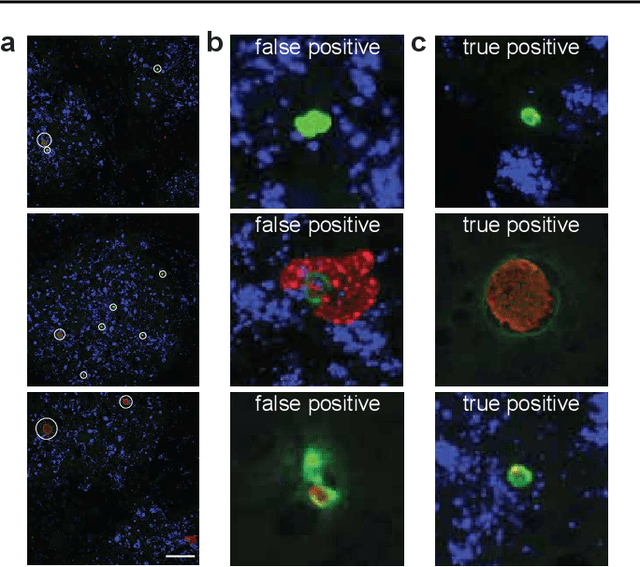
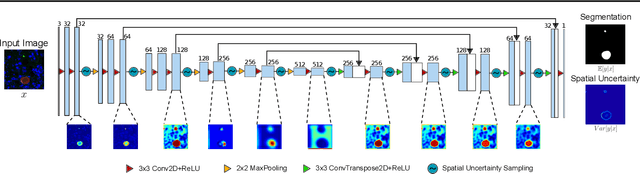
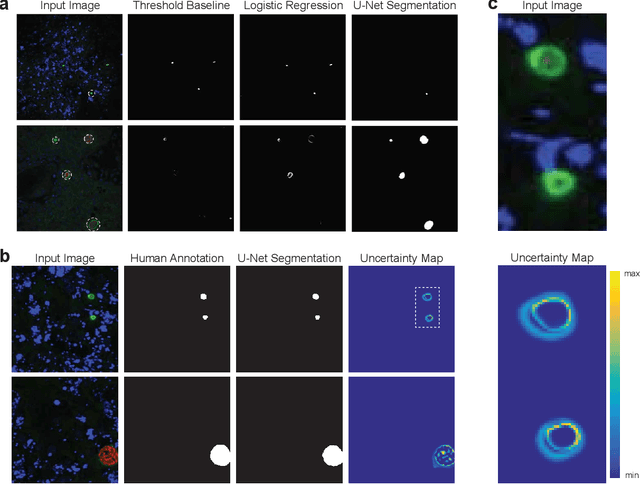
Abstract:Global eradication of malaria depends on the development of drugs effective against the silent, yet obligate liver stage of the disease. The gold standard in drug development remains microscopic imaging of liver stage parasites in in vitro cell culture models. Image analysis presents a major bottleneck in this pipeline since the parasite has significant variability in size, shape, and density in these models. As with other highly variable datasets, traditional segmentation models have poor generalizability as they rely on hand-crafted features; thus, manual annotation of liver stage malaria images remains standard. To address this need, we develop a convolutional neural network architecture that utilizes spatial dropout sampling for parasite segmentation and epistemic uncertainty estimation in images of liver stage malaria. Our pipeline produces high-precision segmentations nearly identical to expert annotations, generalizes well on a diverse dataset of liver stage malaria parasites, and promotes independence between learned feature maps to model the uncertainty of generated predictions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge