Tianyu Ma
UniverSeg: Universal Medical Image Segmentation
Apr 12, 2023



Abstract:While deep learning models have become the predominant method for medical image segmentation, they are typically not capable of generalizing to unseen segmentation tasks involving new anatomies, image modalities, or labels. Given a new segmentation task, researchers generally have to train or fine-tune models, which is time-consuming and poses a substantial barrier for clinical researchers, who often lack the resources and expertise to train neural networks. We present UniverSeg, a method for solving unseen medical segmentation tasks without additional training. Given a query image and example set of image-label pairs that define a new segmentation task, UniverSeg employs a new Cross-Block mechanism to produce accurate segmentation maps without the need for additional training. To achieve generalization to new tasks, we have gathered and standardized a collection of 53 open-access medical segmentation datasets with over 22,000 scans, which we refer to as MegaMedical. We used this collection to train UniverSeg on a diverse set of anatomies and imaging modalities. We demonstrate that UniverSeg substantially outperforms several related methods on unseen tasks, and thoroughly analyze and draw insights about important aspects of the proposed system. The UniverSeg source code and model weights are freely available at https://universeg.csail.mit.edu
Label conditioned segmentation
Mar 17, 2022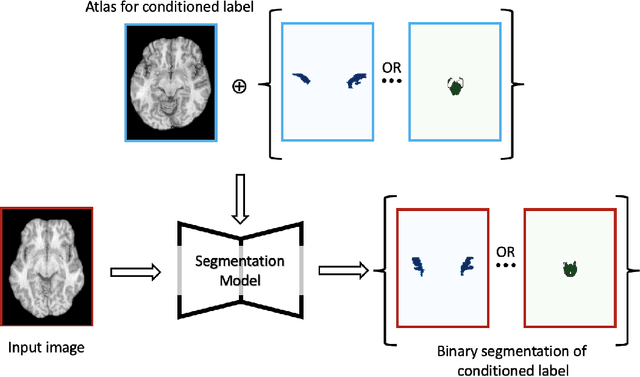
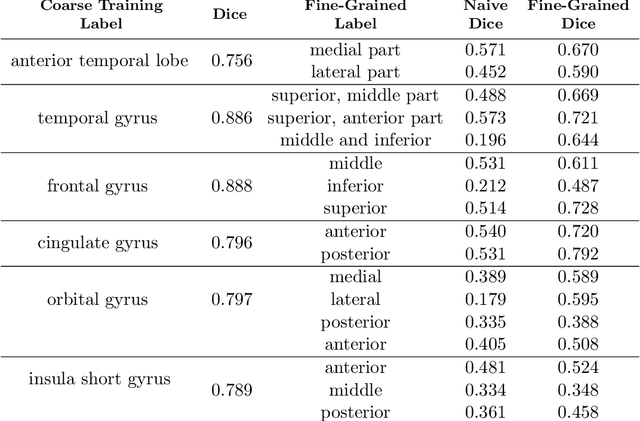
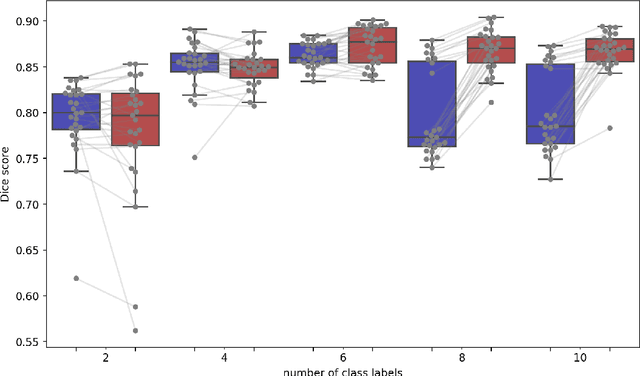
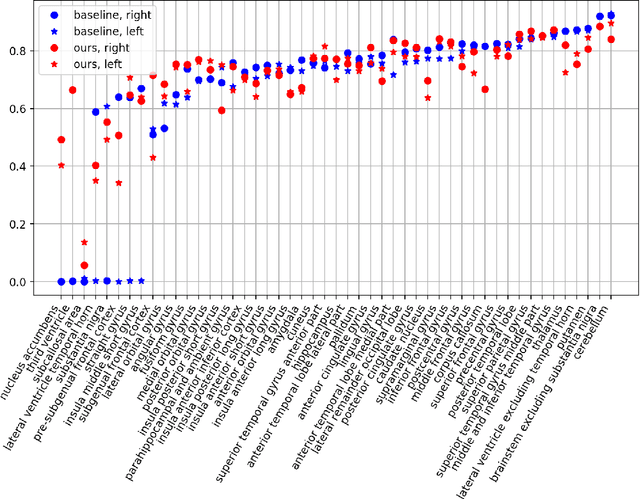
Abstract:Semantic segmentation is an important task in computer vision that is often tackled with convolutional neural networks (CNNs). A CNN learns to produce pixel-level predictions through training on pairs of images and their corresponding ground-truth segmentation labels. For segmentation tasks with multiple classes, the standard approach is to use a network that computes a multi-channel probabilistic segmentation map, with each channel representing one class. In applications where the image grid size (e.g., when it is a 3D volume) and/or the number of labels is relatively large, the standard (baseline) approach can become prohibitively expensive for our computational resources. In this paper, we propose a simple yet effective method to address this challenge. In our approach, the segmentation network produces a single-channel output, while being conditioned on a single class label, which determines the output class of the network. Our method, called label conditioned segmentation (LCS), can be used to segment images with a very large number of classes, which might be infeasible for the baseline approach. We also demonstrate in the experiments that label conditioning can improve the accuracy of a given backbone architecture, likely, thanks to its parameter efficiency. Finally, as we show in our results, an LCS model can produce previously unseen fine-grained labels during inference time, when only coarse labels were available during training. We provide all of our code here: https://github.com/tym002/Label-conditioned-segmentation
Hyper-Convolutions via Implicit Kernels for Medical Imaging
Feb 06, 2022



Abstract:The convolutional neural network (CNN) is one of the most commonly used architectures for computer vision tasks. The key building block of a CNN is the convolutional kernel that aggregates information from the pixel neighborhood and shares weights across all pixels. A standard CNN's capacity, and thus its performance, is directly related to the number of learnable kernel weights, which is determined by the number of channels and the kernel size (support). In this paper, we present the \textit{hyper-convolution}, a novel building block that implicitly encodes the convolutional kernel using spatial coordinates. Hyper-convolutions decouple kernel size from the total number of learnable parameters, enabling a more flexible architecture design. We demonstrate in our experiments that replacing regular convolutions with hyper-convolutions can improve performance with less parameters, and increase robustness against noise. We provide our code here: \emph{https://github.com/tym002/Hyper-Convolution}
Hyper-Convolution Networks for Biomedical Image Segmentation
May 21, 2021



Abstract:The convolution operation is a central building block of neural network architectures widely used in computer vision. The size of the convolution kernels determines both the expressiveness of convolutional neural networks (CNN), as well as the number of learnable parameters. Increasing the network capacity to capture rich pixel relationships requires increasing the number of learnable parameters, often leading to overfitting and/or lack of robustness. In this paper, we propose a powerful novel building block, the hyper-convolution, which implicitly represents the convolution kernel as a function of kernel coordinates. Hyper-convolutions enable decoupling the kernel size, and hence its receptive field, from the number of learnable parameters. In our experiments, focused on challenging biomedical image segmentation tasks, we demonstrate that replacing regular convolutions with hyper-convolutions leads to more efficient architectures that achieve improved accuracy. Our analysis also shows that learned hyper-convolutions are naturally regularized, which can offer better generalization performance. We believe that hyper-convolutions can be a powerful building block in future neural network architectures solving computer vision tasks.
Ensembling Low Precision Models for Binary Biomedical Image Segmentation
Oct 16, 2020



Abstract:Segmentation of anatomical regions of interest such as vessels or small lesions in medical images is still a difficult problem that is often tackled with manual input by an expert. One of the major challenges for this task is that the appearance of foreground (positive) regions can be similar to background (negative) regions. As a result, many automatic segmentation algorithms tend to exhibit asymmetric errors, typically producing more false positives than false negatives. In this paper, we aim to leverage this asymmetry and train a diverse ensemble of models with very high recall, while sacrificing their precision. Our core idea is straightforward: A diverse ensemble of low precision and high recall models are likely to make different false positive errors (classifying background as foreground in different parts of the image), but the true positives will tend to be consistent. Thus, in aggregate the false positive errors will cancel out, yielding high performance for the ensemble. Our strategy is general and can be applied with any segmentation model. In three different applications (carotid artery segmentation in a neck CT angiography, myocardium segmentation in a cardiovascular MRI and multiple sclerosis lesion segmentation in a brain MRI), we show how the proposed approach can significantly boost the performance of a baseline segmentation method.
Volumetric landmark detection with a multi-scale shift equivariant neural network
Mar 03, 2020



Abstract:Deep neural networks yield promising results in a wide range of computer vision applications, including landmark detection. A major challenge for accurate anatomical landmark detection in volumetric images such as clinical CT scans is that large-scale data often constrain the capacity of the employed neural network architecture due to GPU memory limitations, which in turn can limit the precision of the output. We propose a multi-scale, end-to-end deep learning method that achieves fast and memory-efficient landmark detection in 3D images. Our architecture consists of blocks of shift-equivariant networks, each of which performs landmark detection at a different spatial scale. These blocks are connected from coarse to fine-scale, with differentiable resampling layers, so that all levels can be trained together. We also present a noise injection strategy that increases the robustness of the model and allows us to quantify uncertainty at test time. We evaluate our method for carotid artery bifurcations detection on 263 CT volumes and achieve a better than state-of-the-art accuracy with mean Euclidean distance error of 2.81mm.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge