Victor Ion Butoi
VoxelPrompt: A Vision-Language Agent for Grounded Medical Image Analysis
Oct 10, 2024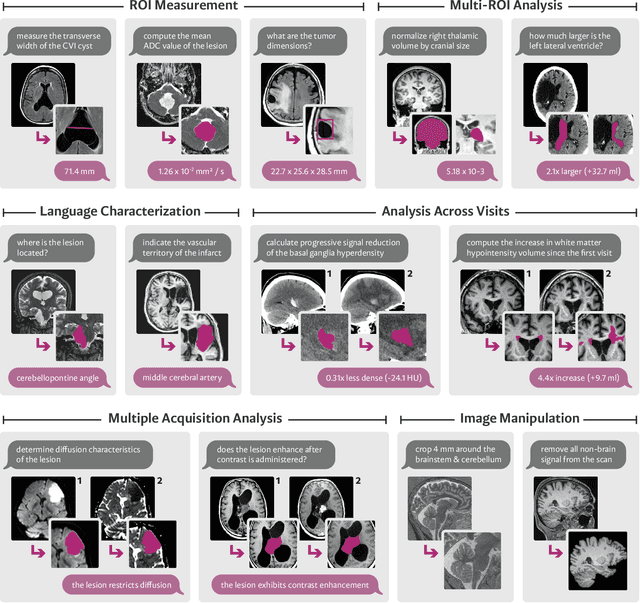
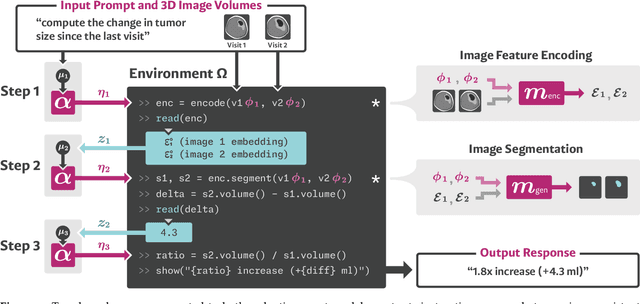
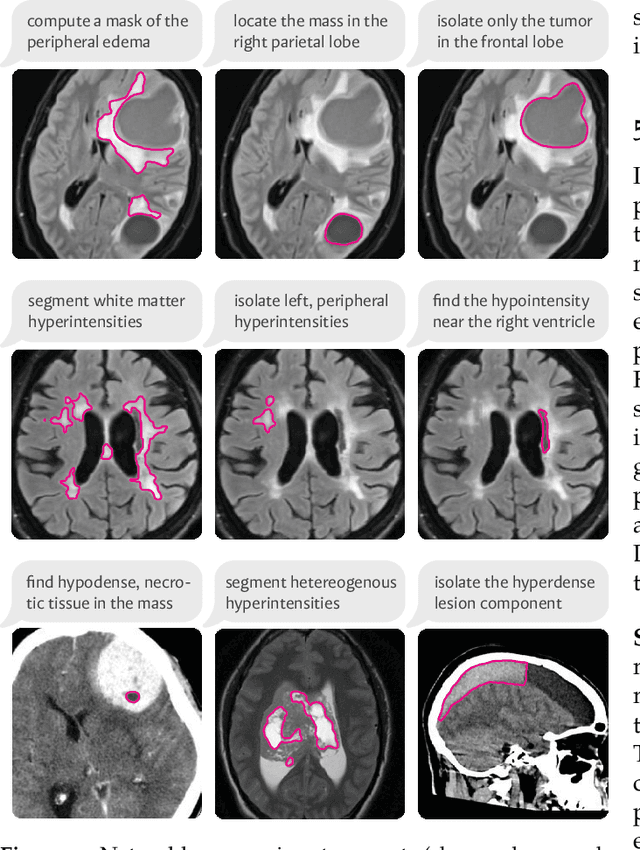
Abstract:We present VoxelPrompt, an agent-driven vision-language framework that tackles diverse radiological tasks through joint modeling of natural language, image volumes, and analytical metrics. VoxelPrompt is multi-modal and versatile, leveraging the flexibility of language interaction while providing quantitatively grounded image analysis. Given a variable number of 3D medical volumes, such as MRI and CT scans, VoxelPrompt employs a language agent that iteratively predicts executable instructions to solve a task specified by an input prompt. These instructions communicate with a vision network to encode image features and generate volumetric outputs (e.g., segmentations). VoxelPrompt interprets the results of intermediate instructions and plans further actions to compute discrete measures (e.g., tumor growth across a series of scans) and present relevant outputs to the user. We evaluate this framework in a sandbox of diverse neuroimaging tasks, and we show that the single VoxelPrompt model can delineate hundreds of anatomical and pathological features, measure many complex morphological properties, and perform open-language analysis of lesion characteristics. VoxelPrompt carries out these objectives with accuracy similar to that of fine-tuned, single-task models for segmentation and visual question-answering, while facilitating a much larger range of tasks. Therefore, by supporting accurate image processing with language interaction, VoxelPrompt provides comprehensive utility for numerous imaging tasks that traditionally require specialized models to address.
ConMe: Rethinking Evaluation of Compositional Reasoning for Modern VLMs
Jun 12, 2024



Abstract:Compositional Reasoning (CR) entails grasping the significance of attributes, relations, and word order. Recent Vision-Language Models (VLMs), comprising a visual encoder and a Large Language Model (LLM) decoder, have demonstrated remarkable proficiency in such reasoning tasks. This prompts a crucial question: have VLMs effectively tackled the CR challenge? We conjecture that existing CR benchmarks may not adequately push the boundaries of modern VLMs due to the reliance on an LLM-only negative text generation pipeline. Consequently, the negatives produced either appear as outliers from the natural language distribution learned by VLMs' LLM decoders or as improbable within the corresponding image context. To address these limitations, we introduce ConMe -- a compositional reasoning benchmark and a novel data generation pipeline leveraging VLMs to produce `hard CR Q&A'. Through a new concept of VLMs conversing with each other to collaboratively expose their weaknesses, our pipeline autonomously generates, evaluates, and selects challenging compositional reasoning questions, establishing a robust CR benchmark, also subsequently validated manually. Our benchmark provokes a noteworthy, up to 33%, decrease in CR performance compared to preceding benchmarks, reinstating the CR challenge even for state-of-the-art VLMs.
Generative Active Learning for the Search of Small-molecule Protein Binders
May 02, 2024



Abstract:Despite substantial progress in machine learning for scientific discovery in recent years, truly de novo design of small molecules which exhibit a property of interest remains a significant challenge. We introduce LambdaZero, a generative active learning approach to search for synthesizable molecules. Powered by deep reinforcement learning, LambdaZero learns to search over the vast space of molecules to discover candidates with a desired property. We apply LambdaZero with molecular docking to design novel small molecules that inhibit the enzyme soluble Epoxide Hydrolase 2 (sEH), while enforcing constraints on synthesizability and drug-likeliness. LambdaZero provides an exponential speedup in terms of the number of calls to the expensive molecular docking oracle, and LambdaZero de novo designed molecules reach docking scores that would otherwise require the virtual screening of a hundred billion molecules. Importantly, LambdaZero discovers novel scaffolds of synthesizable, drug-like inhibitors for sEH. In in vitro experimental validation, a series of ligands from a generated quinazoline-based scaffold were synthesized, and the lead inhibitor N-(4,6-di(pyrrolidin-1-yl)quinazolin-2-yl)-N-methylbenzamide (UM0152893) displayed sub-micromolar enzyme inhibition of sEH.
Empirical Analysis of a Segmentation Foundation Model in Prostate Imaging
Jul 06, 2023

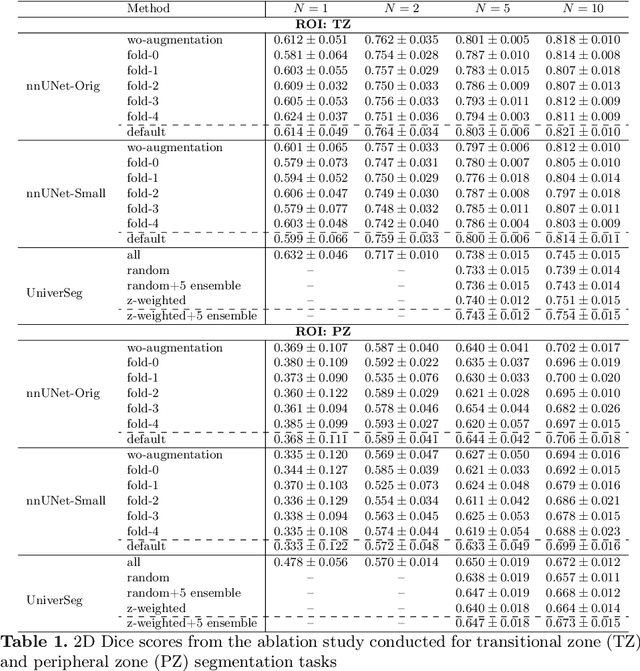
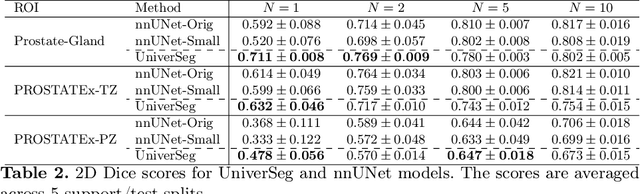
Abstract:Most state-of-the-art techniques for medical image segmentation rely on deep-learning models. These models, however, are often trained on narrowly-defined tasks in a supervised fashion, which requires expensive labeled datasets. Recent advances in several machine learning domains, such as natural language generation have demonstrated the feasibility and utility of building foundation models that can be customized for various downstream tasks with little to no labeled data. This likely represents a paradigm shift for medical imaging, where we expect that foundation models may shape the future of the field. In this paper, we consider a recently developed foundation model for medical image segmentation, UniverSeg. We conduct an empirical evaluation study in the context of prostate imaging and compare it against the conventional approach of training a task-specific segmentation model. Our results and discussion highlight several important factors that will likely be important in the development and adoption of foundation models for medical image segmentation.
UniverSeg: Universal Medical Image Segmentation
Apr 12, 2023



Abstract:While deep learning models have become the predominant method for medical image segmentation, they are typically not capable of generalizing to unseen segmentation tasks involving new anatomies, image modalities, or labels. Given a new segmentation task, researchers generally have to train or fine-tune models, which is time-consuming and poses a substantial barrier for clinical researchers, who often lack the resources and expertise to train neural networks. We present UniverSeg, a method for solving unseen medical segmentation tasks without additional training. Given a query image and example set of image-label pairs that define a new segmentation task, UniverSeg employs a new Cross-Block mechanism to produce accurate segmentation maps without the need for additional training. To achieve generalization to new tasks, we have gathered and standardized a collection of 53 open-access medical segmentation datasets with over 22,000 scans, which we refer to as MegaMedical. We used this collection to train UniverSeg on a diverse set of anatomies and imaging modalities. We demonstrate that UniverSeg substantially outperforms several related methods on unseen tasks, and thoroughly analyze and draw insights about important aspects of the proposed system. The UniverSeg source code and model weights are freely available at https://universeg.csail.mit.edu
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge