Heejong Kim
AI-Based Detection of In-Treatment Changes from Prostate MR-Linac Images
Feb 04, 2026Abstract:Purpose: To investigate whether routinely acquired longitudinal MR-Linac images can be leveraged to characterize treatment-induced changes during radiotherapy, particularly subtle inter-fraction changes over short intervals (average of 2 days). Materials and Methods: This retrospective study included a series of 0.35T MR-Linac images from 761 patients. An artificial intelligence (deep learning) model was used to characterize treatment-induced changes by predicting the temporal order of paired images. The model was first trained with the images from the first and the last fractions (F1-FL), then with all pairs (All-pairs). Model performance was assessed using quantitative metrics (accuracy and AUC), compared to a radiologist's performance, and qualitative analyses - the saliency map evaluation to investigate affected anatomical regions. Input ablation experiments were performed to identify the anatomical regions altered by radiotherapy. The radiologist conducted an additional task on partial images reconstructed by saliency map regions, reporting observations as well. Quantitative image analysis was conducted to investigate the results from the model and the radiologist. Results: The F1-FL model yielded near-perfect performance (AUC of 0.99), significantly outperforming the radiologist. The All-pairs model yielded an AUC of 0.97. This performance reflects therapy-induced changes, supported by the performance correlation to fraction intervals, ablation tests and expert's interpretation. Primary regions driving the predictions were prostate, bladder, and pubic symphysis. Conclusion: The model accurately predicts temporal order of MR-Linac fractions and detects radiation-induced changes over one or a few days, including prostate and adjacent organ alterations confirmed by experts. This underscores MR-Linac's potential for advanced image analysis beyond image guidance.
Effective Segmentation of Post-Treatment Gliomas Using Simple Approaches: Artificial Sequence Generation and Ensemble Models
Sep 12, 2024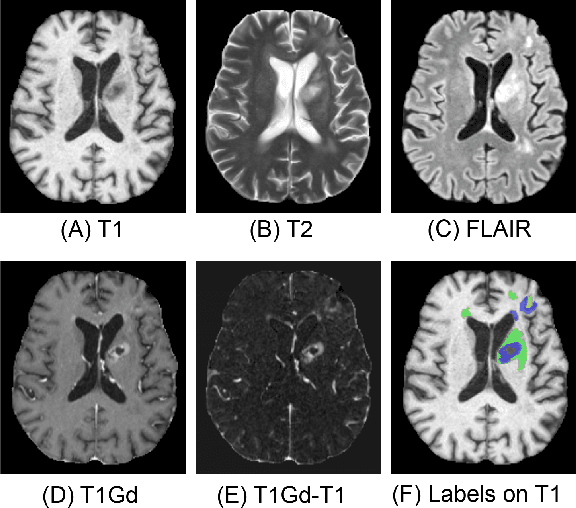
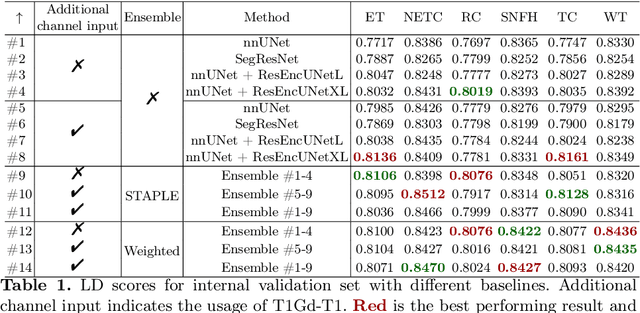
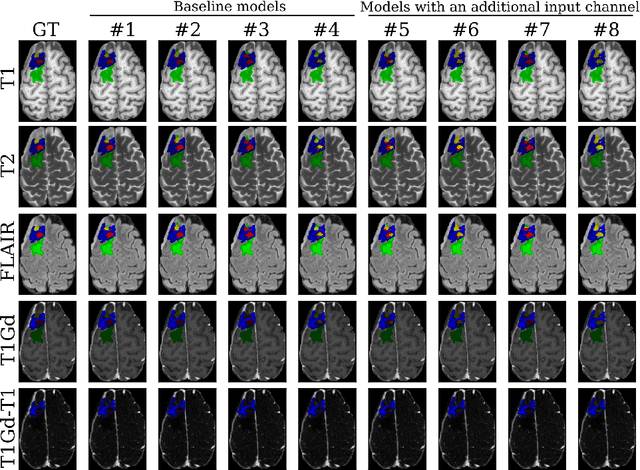
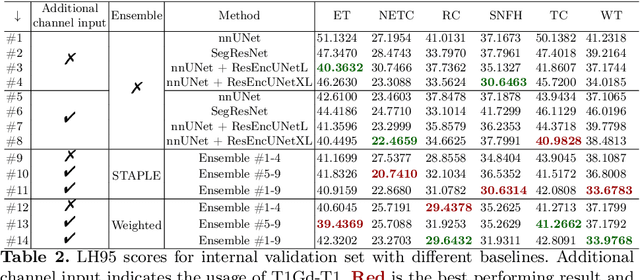
Abstract:Segmentation is a crucial task in the medical imaging field and is often an important primary step or even a prerequisite to the analysis of medical volumes. Yet treatments such as surgery complicate the accurate delineation of regions of interest. The BraTS Post-Treatment 2024 Challenge published the first public dataset for post-surgery glioma segmentation and addresses the aforementioned issue by fostering the development of automated segmentation tools for glioma in MRI data. In this effort, we propose two straightforward approaches to enhance the segmentation performances of deep learning-based methodologies. First, we incorporate an additional input based on a simple linear combination of the available MRI sequences input, which highlights enhancing tumors. Second, we employ various ensembling methods to weigh the contribution of a battery of models. Our results demonstrate that these approaches significantly improve segmentation performance compared to baseline models, underscoring the effectiveness of these simple approaches in improving medical image segmentation tasks.
Adapting to Shifting Correlations with Unlabeled Data Calibration
Sep 09, 2024



Abstract:Distribution shifts between sites can seriously degrade model performance since models are prone to exploiting unstable correlations. Thus, many methods try to find features that are stable across sites and discard unstable features. However, unstable features might have complementary information that, if used appropriately, could increase accuracy. More recent methods try to adapt to unstable features at the new sites to achieve higher accuracy. However, they make unrealistic assumptions or fail to scale to multiple confounding features. We propose Generalized Prevalence Adjustment (GPA for short), a flexible method that adjusts model predictions to the shifting correlations between prediction target and confounders to safely exploit unstable features. GPA can infer the interaction between target and confounders in new sites using unlabeled samples from those sites. We evaluate GPA on several real and synthetic datasets, and show that it outperforms competitive baselines.
Knockout: A simple way to handle missing inputs
Jun 03, 2024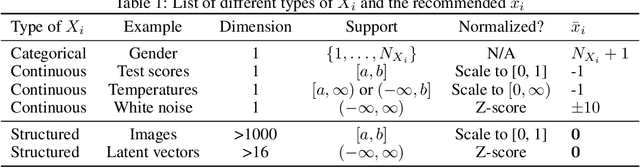

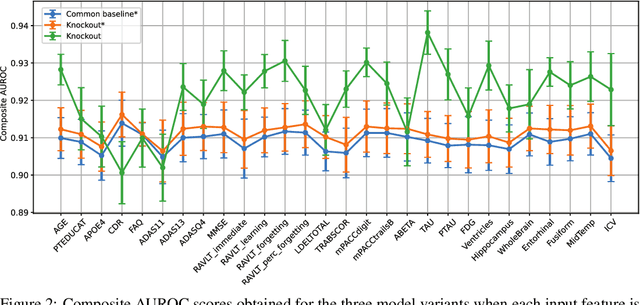
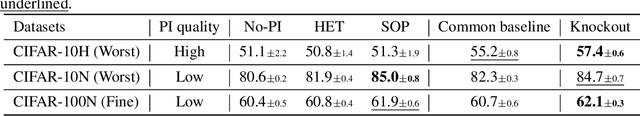
Abstract:Deep learning models can extract predictive and actionable information from complex inputs. The richer the inputs, the better these models usually perform. However, models that leverage rich inputs (e.g., multi-modality) can be difficult to deploy widely, because some inputs may be missing at inference. Current popular solutions to this problem include marginalization, imputation, and training multiple models. Marginalization can obtain calibrated predictions but it is computationally costly and therefore only feasible for low dimensional inputs. Imputation may result in inaccurate predictions because it employs point estimates for missing variables and does not work well for high dimensional inputs (e.g., images). Training multiple models whereby each model takes different subsets of inputs can work well but requires knowing missing input patterns in advance. Furthermore, training and retaining multiple models can be costly. We propose an efficient way to learn both the conditional distribution using full inputs and the marginal distributions. Our method, Knockout, randomly replaces input features with appropriate placeholder values during training. We provide a theoretical justification of Knockout and show that it can be viewed as an implicit marginalization strategy. We evaluate Knockout in a wide range of simulations and real-world datasets and show that it can offer strong empirical performance.
BrainMorph: A Foundational Keypoint Model for Robust and Flexible Brain MRI Registration
May 22, 2024Abstract:We present a keypoint-based foundation model for general purpose brain MRI registration, based on the recently-proposed KeyMorph framework. Our model, called BrainMorph, serves as a tool that supports multi-modal, pairwise, and scalable groupwise registration. BrainMorph is trained on a massive dataset of over 100,000 3D volumes, skull-stripped and non-skull-stripped, from nearly 16,000 unique healthy and diseased subjects. BrainMorph is robust to large misalignments, interpretable via interrogating automatically-extracted keypoints, and enables rapid and controllable generation of many plausible transformations with different alignment types and different degrees of nonlinearity at test-time. We demonstrate the superiority of BrainMorph in solving 3D rigid, affine, and nonlinear registration on a variety of multi-modal brain MRI scans of healthy and diseased subjects, in both the pairwise and groupwise setting. In particular, we show registration accuracy and speeds that surpass current state-of-the-art methods, especially in the context of large initial misalignments and large group settings. All code and models are available at https://github.com/alanqrwang/brainmorph.
Robust Learning via Conditional Prevalence Adjustment
Oct 24, 2023Abstract:Healthcare data often come from multiple sites in which the correlations between confounding variables can vary widely. If deep learning models exploit these unstable correlations, they might fail catastrophically in unseen sites. Although many methods have been proposed to tackle unstable correlations, each has its limitations. For example, adversarial training forces models to completely ignore unstable correlations, but doing so may lead to poor predictive performance. Other methods (e.g. Invariant risk minimization [4]) try to learn domain-invariant representations that rely only on stable associations by assuming a causal data-generating process (input X causes class label Y ). Thus, they may be ineffective for anti-causal tasks (Y causes X), which are common in computer vision. We propose a method called CoPA (Conditional Prevalence-Adjustment) for anti-causal tasks. CoPA assumes that (1) generation mechanism is stable, i.e. label Y and confounding variable(s) Z generate X, and (2) the unstable conditional prevalence in each site E fully accounts for the unstable correlations between X and Y . Our crucial observation is that confounding variables are routinely recorded in healthcare settings and the prevalence can be readily estimated, for example, from a set of (Y, Z) samples (no need for corresponding samples of X). CoPA can work even if there is a single training site, a scenario which is often overlooked by existing methods. Our experiments on synthetic and real data show CoPA beating competitive baselines.
A Framework for Interpretability in Machine Learning for Medical Imaging
Oct 02, 2023

Abstract:Interpretability for machine learning models in medical imaging (MLMI) is an important direction of research. However, there is a general sense of murkiness in what interpretability means. Why does the need for interpretability in MLMI arise? What goals does one actually seek to address when interpretability is needed? To answer these questions, we identify a need to formalize the goals and elements of interpretability in MLMI. By reasoning about real-world tasks and goals common in both medical image analysis and its intersection with machine learning, we identify four core elements of interpretability: localization, visual recognizability, physical attribution, and transparency. Overall, this paper formalizes interpretability needs in the context of medical imaging, and our applied perspective clarifies concrete MLMI-specific goals and considerations in order to guide method design and improve real-world usage. Our goal is to provide practical and didactic information for model designers and practitioners, inspire developers of models in the medical imaging field to reason more deeply about what interpretability is achieving, and suggest future directions of interpretability research.
Empirical Analysis of a Segmentation Foundation Model in Prostate Imaging
Jul 06, 2023

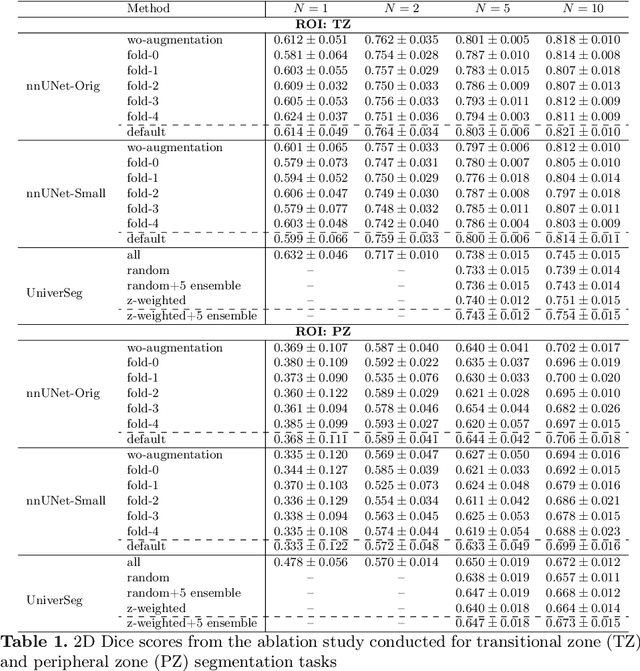
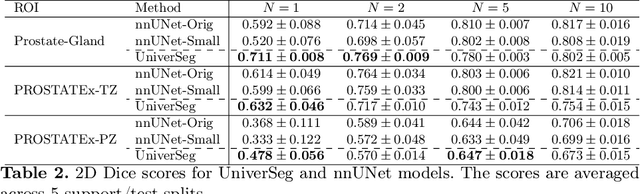
Abstract:Most state-of-the-art techniques for medical image segmentation rely on deep-learning models. These models, however, are often trained on narrowly-defined tasks in a supervised fashion, which requires expensive labeled datasets. Recent advances in several machine learning domains, such as natural language generation have demonstrated the feasibility and utility of building foundation models that can be customized for various downstream tasks with little to no labeled data. This likely represents a paradigm shift for medical imaging, where we expect that foundation models may shape the future of the field. In this paper, we consider a recently developed foundation model for medical image segmentation, UniverSeg. We conduct an empirical evaluation study in the context of prostate imaging and compare it against the conventional approach of training a task-specific segmentation model. Our results and discussion highlight several important factors that will likely be important in the development and adoption of foundation models for medical image segmentation.
Learning to Compare Longitudinal Images
Apr 16, 2023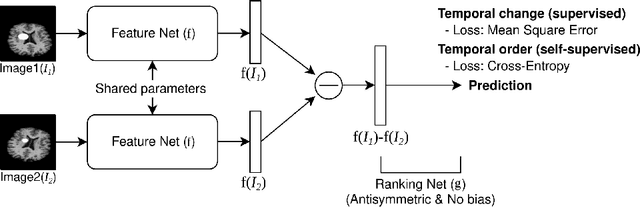

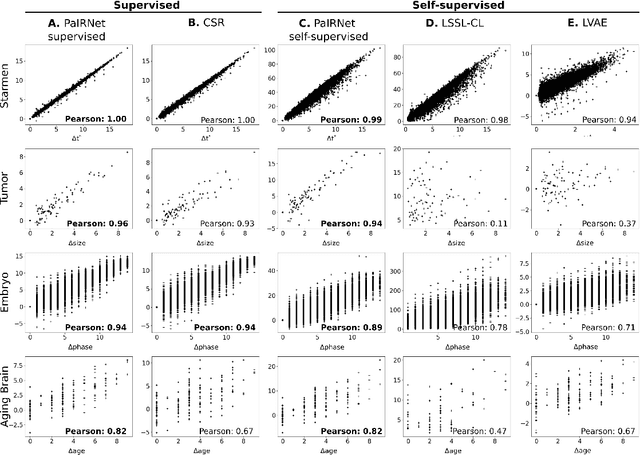

Abstract:Longitudinal studies, where a series of images from the same set of individuals are acquired at different time-points, represent a popular technique for studying and characterizing temporal dynamics in biomedical applications. The classical approach for longitudinal comparison involves normalizing for nuisance variations, such as image orientation or contrast differences, via pre-processing. Statistical analysis is, in turn, conducted to detect changes of interest, either at the individual or population level. This classical approach can suffer from pre-processing issues and limitations of the statistical modeling. For example, normalizing for nuisance variation might be hard in settings where there are a lot of idiosyncratic changes. In this paper, we present a simple machine learning-based approach that can alleviate these issues. In our approach, we train a deep learning model (called PaIRNet, for Pairwise Image Ranking Network) to compare pairs of longitudinal images, with or without supervision. In the self-supervised setup, for instance, the model is trained to temporally order the images, which requires learning to recognize time-irreversible changes. Our results from four datasets demonstrate that PaIRNet can be very effective in localizing and quantifying meaningful longitudinal changes while discounting nuisance variation. Our code is available at \url{https://github.com/heejong-kim/learning-to-compare-longitudinal-images.git}
Q-space Conditioned Translation Networks for Directional Synthesis of Diffusion Weighted Images from Multi-modal Structural MRI
Jun 24, 2021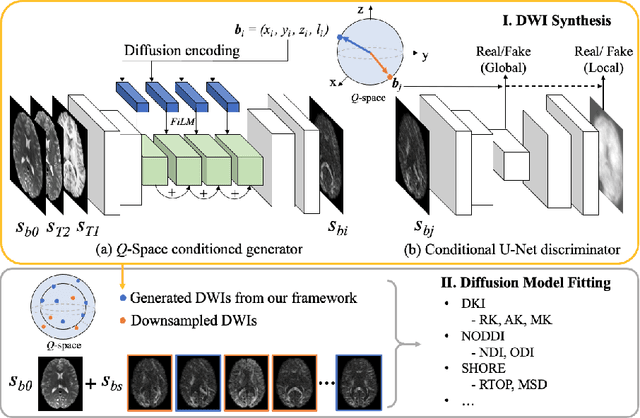
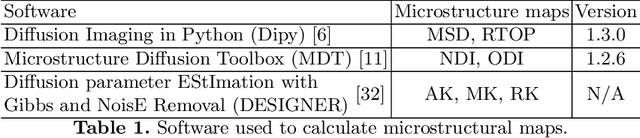
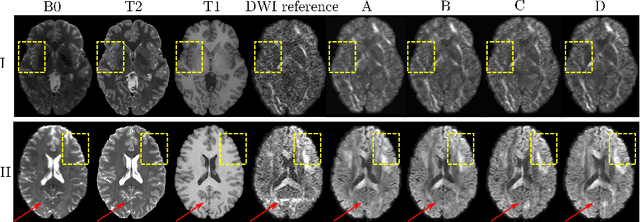
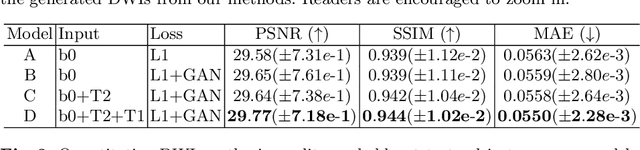
Abstract:Current deep learning approaches for diffusion MRI modeling circumvent the need for densely-sampled diffusion-weighted images (DWIs) by directly predicting microstructural indices from sparsely-sampled DWIs. However, they implicitly make unrealistic assumptions of static $q$-space sampling during training and reconstruction. Further, such approaches can restrict downstream usage of variably sampled DWIs for usages including the estimation of microstructural indices or tractography. We propose a generative adversarial translation framework for high-quality DWI synthesis with arbitrary $q$-space sampling given commonly acquired structural images (e.g., B0, T1, T2). Our translation network linearly modulates its internal representations conditioned on continuous $q$-space information, thus removing the need for fixed sampling schemes. Moreover, this approach enables downstream estimation of high-quality microstructural maps from arbitrarily subsampled DWIs, which may be particularly important in cases with sparsely sampled DWIs. Across several recent methodologies, the proposed approach yields improved DWI synthesis accuracy and fidelity with enhanced downstream utility as quantified by the accuracy of scalar microstructure indices estimated from the synthesized images. Code is available at https://github.com/mengweiren/q-space-conditioned-dwi-synthesis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge