Guido Gerig
Equivariant spatio-hemispherical networks for diffusion MRI deconvolution
Nov 18, 2024



Abstract:Each voxel in a diffusion MRI (dMRI) image contains a spherical signal corresponding to the direction and strength of water diffusion in the brain. This paper advances the analysis of such spatio-spherical data by developing convolutional network layers that are equivariant to the $\mathbf{E(3) \times SO(3)}$ group and account for the physical symmetries of dMRI including rotations, translations, and reflections of space alongside voxel-wise rotations. Further, neuronal fibers are typically antipodally symmetric, a fact we leverage to construct highly efficient spatio-hemispherical graph convolutions to accelerate the analysis of high-dimensional dMRI data. In the context of sparse spherical fiber deconvolution to recover white matter microstructure, our proposed equivariant network layers yield substantial performance and efficiency gains, leading to better and more practical resolution of crossing neuronal fibers and fiber tractography. These gains are experimentally consistent across both simulation and in vivo human datasets.
Relightful Harmonization: Lighting-aware Portrait Background Replacement
Dec 11, 2023



Abstract:Portrait harmonization aims to composite a subject into a new background, adjusting its lighting and color to ensure harmony with the background scene. Existing harmonization techniques often only focus on adjusting the global color and brightness of the foreground and ignore crucial illumination cues from the background such as apparent lighting direction, leading to unrealistic compositions. We introduce Relightful Harmonization, a lighting-aware diffusion model designed to seamlessly harmonize sophisticated lighting effect for the foreground portrait using any background image. Our approach unfolds in three stages. First, we introduce a lighting representation module that allows our diffusion model to encode lighting information from target image background. Second, we introduce an alignment network that aligns lighting features learned from image background with lighting features learned from panorama environment maps, which is a complete representation for scene illumination. Last, to further boost the photorealism of the proposed method, we introduce a novel data simulation pipeline that generates synthetic training pairs from a diverse range of natural images, which are used to refine the model. Our method outperforms existing benchmarks in visual fidelity and lighting coherence, showing superior generalization in real-world testing scenarios, highlighting its versatility and practicality.
Self-supervised OCT Image Denoising with Slice-to-Slice Registration and Reconstruction
Dec 08, 2023



Abstract:Strong speckle noise is inherent to optical coherence tomography (OCT) imaging and represents a significant obstacle for accurate quantitative analysis of retinal structures which is key for advances in clinical diagnosis and monitoring of disease. Learning-based self-supervised methods for structure-preserving noise reduction have demonstrated superior performance over traditional methods but face unique challenges in OCT imaging. The high correlation of voxels generated by coherent A-scan beams undermines the efficacy of self-supervised learning methods as it violates the assumption of independent pixel noise. We conduct experiments demonstrating limitations of existing models due to this independence assumption. We then introduce a new end-to-end self-supervised learning framework specifically tailored for OCT image denoising, integrating slice-by-slice training and registration modules into one network. An extensive ablation study is conducted for the proposed approach. Comparison to previously published self-supervised denoising models demonstrates improved performance of the proposed framework, potentially serving as a preprocessing step towards superior segmentation performance and quantitative analysis.
Keypoint-Augmented Self-Supervised Learning for Medical Image Segmentation with Limited Annotation
Oct 18, 2023



Abstract:Pretraining CNN models (i.e., UNet) through self-supervision has become a powerful approach to facilitate medical image segmentation under low annotation regimes. Recent contrastive learning methods encourage similar global representations when the same image undergoes different transformations, or enforce invariance across different image/patch features that are intrinsically correlated. However, CNN-extracted global and local features are limited in capturing long-range spatial dependencies that are essential in biological anatomy. To this end, we present a keypoint-augmented fusion layer that extracts representations preserving both short- and long-range self-attention. In particular, we augment the CNN feature map at multiple scales by incorporating an additional input that learns long-range spatial self-attention among localized keypoint features. Further, we introduce both global and local self-supervised pretraining for the framework. At the global scale, we obtain global representations from both the bottleneck of the UNet, and by aggregating multiscale keypoint features. These global features are subsequently regularized through image-level contrastive objectives. At the local scale, we define a distance-based criterion to first establish correspondences among keypoints and encourage similarity between their features. Through extensive experiments on both MRI and CT segmentation tasks, we demonstrate the architectural advantages of our proposed method in comparison to both CNN and Transformer-based UNets, when all architectures are trained with randomly initialized weights. With our proposed pretraining strategy, our method further outperforms existing SSL methods by producing more robust self-attention and achieving state-of-the-art segmentation results. The code is available at https://github.com/zshyang/kaf.git.
Microscopy Image Segmentation via Point and Shape Regularized Data Synthesis
Aug 18, 2023Abstract:Current deep learning-based approaches for the segmentation of microscopy images heavily rely on large amount of training data with dense annotation, which is highly costly and laborious in practice. Compared to full annotation where the complete contour of objects is depicted, point annotations, specifically object centroids, are much easier to acquire and still provide crucial information about the objects for subsequent segmentation. In this paper, we assume access to point annotations only during training and develop a unified pipeline for microscopy image segmentation using synthetically generated training data. Our framework includes three stages: (1) it takes point annotations and samples a pseudo dense segmentation mask constrained with shape priors; (2) with an image generative model trained in an unpaired manner, it translates the mask to a realistic microscopy image regularized by object level consistency; (3) the pseudo masks along with the synthetic images then constitute a pairwise dataset for training an ad-hoc segmentation model. On the public MoNuSeg dataset, our synthesis pipeline produces more diverse and realistic images than baseline models while maintaining high coherence between input masks and generated images. When using the identical segmentation backbones, the models trained on our synthetic dataset significantly outperform those trained with pseudo-labels or baseline-generated images. Moreover, our framework achieves comparable results to models trained on authentic microscopy images with dense labels, demonstrating its potential as a reliable and highly efficient alternative to labor-intensive manual pixel-wise annotations in microscopy image segmentation. The code is available.
$E \times SO$-Equivariant Networks for Spherical Deconvolution in Diffusion MRI
Apr 12, 2023Abstract:We present Roto-Translation Equivariant Spherical Deconvolution (RT-ESD), an $E(3)\times SO(3)$ equivariant framework for sparse deconvolution of volumes where each voxel contains a spherical signal. Such 6D data naturally arises in diffusion MRI (dMRI), a medical imaging modality widely used to measure microstructure and structural connectivity. As each dMRI voxel is typically a mixture of various overlapping structures, there is a need for blind deconvolution to recover crossing anatomical structures such as white matter tracts. Existing dMRI work takes either an iterative or deep learning approach to sparse spherical deconvolution, yet it typically does not account for relationships between neighboring measurements. This work constructs equivariant deep learning layers which respect to symmetries of spatial rotations, reflections, and translations, alongside the symmetries of voxelwise spherical rotations. As a result, RT-ESD improves on previous work across several tasks including fiber recovery on the DiSCo dataset, deconvolution-derived partial volume estimation on real-world \textit{in vivo} human brain dMRI, and improved downstream reconstruction of fiber tractograms on the Tractometer dataset. Our implementation is available at https://github.com/AxelElaldi/e3so3_conv
Image Deblurring with Domain Generalizable Diffusion Models
Dec 04, 2022



Abstract:Diffusion Probabilistic Models (DPMs) have recently been employed for image deblurring. DPMs are trained via a stochastic denoising process that maps Gaussian noise to the high-quality image, conditioned on the concatenated blurry input. Despite their high-quality generated samples, image-conditioned Diffusion Probabilistic Models (icDPM) rely on synthetic pairwise training data (in-domain), with potentially unclear robustness towards real-world unseen images (out-of-domain). In this work, we investigate the generalization ability of icDPMs in deblurring, and propose a simple but effective guidance to significantly alleviate artifacts, and improve the out-of-distribution performance. Particularly, we propose to first extract a multiscale domain-generalizable representation from the input image that removes domain-specific information while preserving the underlying image structure. The representation is then added into the feature maps of the conditional diffusion model as an extra guidance that helps improving the generalization. To benchmark, we focus on out-of-distribution performance by applying a single-dataset trained model to three external and diverse test sets. The effectiveness of the proposed formulation is demonstrated by improvements over the standard icDPM, as well as state-of-the-art performance on perceptual quality and competitive distortion metrics compared to existing methods.
ContraReg: Contrastive Learning of Multi-modality Unsupervised Deformable Image Registration
Jun 27, 2022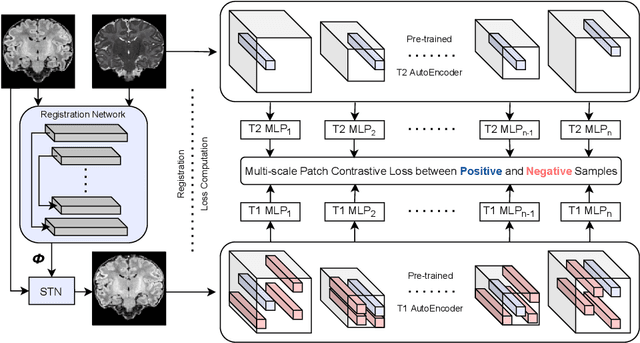
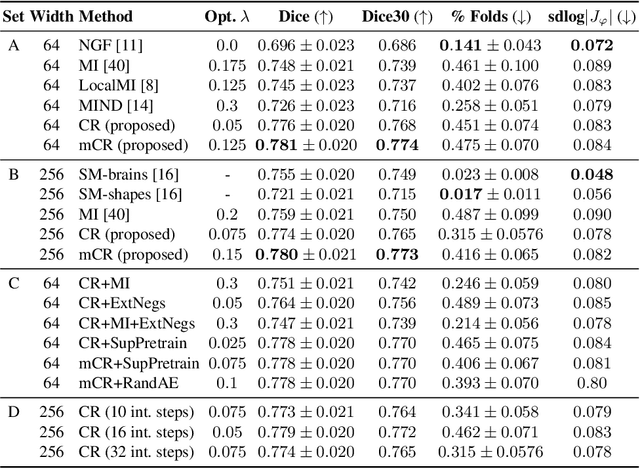
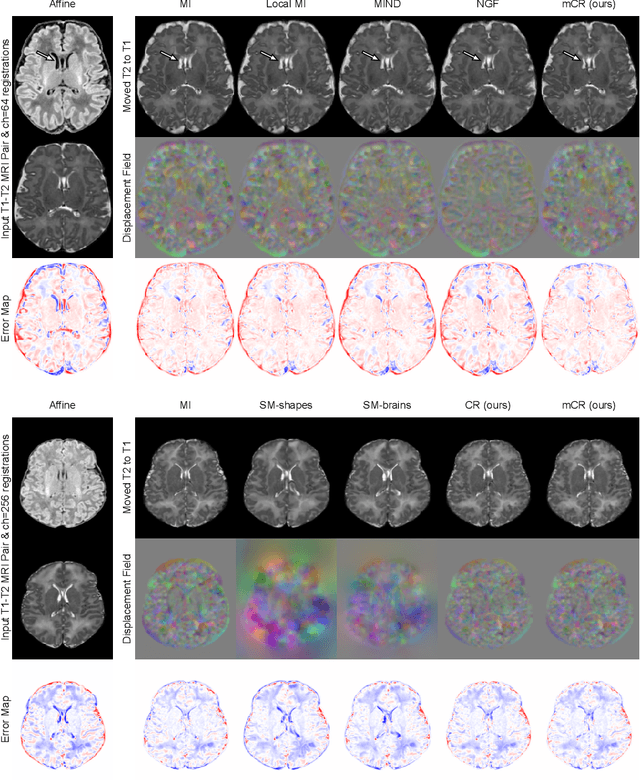
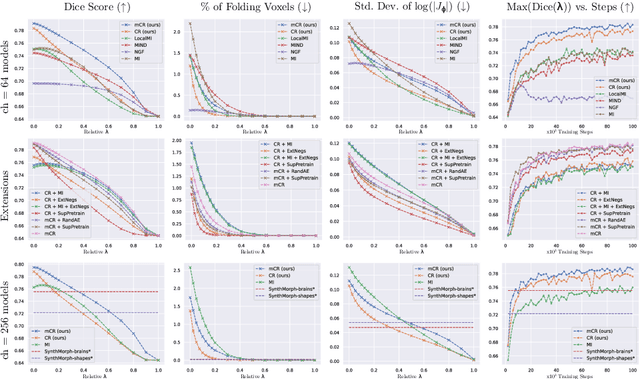
Abstract:Establishing voxelwise semantic correspondence across distinct imaging modalities is a foundational yet formidable computer vision task. Current multi-modality registration techniques maximize hand-crafted inter-domain similarity functions, are limited in modeling nonlinear intensity-relationships and deformations, and may require significant re-engineering or underperform on new tasks, datasets, and domain pairs. This work presents ContraReg, an unsupervised contrastive representation learning approach to multi-modality deformable registration. By projecting learned multi-scale local patch features onto a jointly learned inter-domain embedding space, ContraReg obtains representations useful for non-rigid multi-modality alignment. Experimentally, ContraReg achieves accurate and robust results with smooth and invertible deformations across a series of baselines and ablations on a neonatal T1-T2 brain MRI registration task with all methods validated over a wide range of deformation regularization strengths.
Local Spatiotemporal Representation Learning for Longitudinally-consistent Neuroimage Analysis
Jun 09, 2022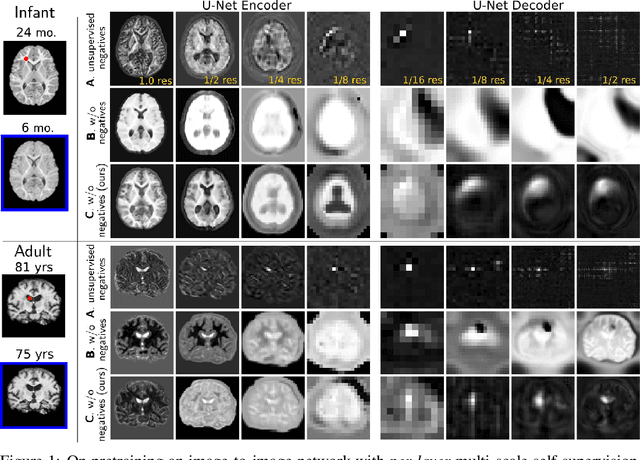
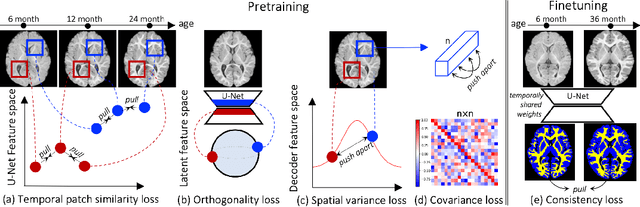

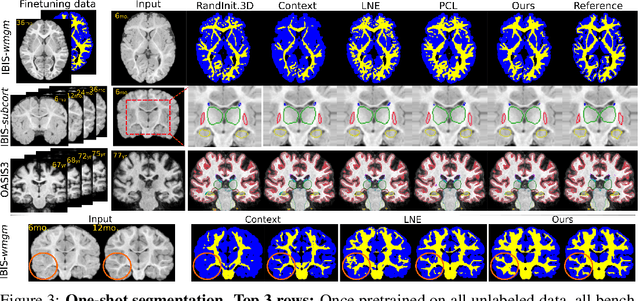
Abstract:Recent self-supervised advances in medical computer vision exploit global and local anatomical self-similarity for pretraining prior to downstream tasks such as segmentation. However, current methods assume i.i.d. image acquisition, which is invalid in clinical study designs where follow-up longitudinal scans track subject-specific temporal changes. Further, existing self-supervised methods for medically-relevant image-to-image architectures exploit only spatial or temporal self-similarity and only do so via a loss applied at a single image-scale, with naive multi-scale spatiotemporal extensions collapsing to degenerate solutions. To these ends, this paper makes two contributions: (1) It presents a local and multi-scale spatiotemporal representation learning method for image-to-image architectures trained on longitudinal images. It exploits the spatiotemporal self-similarity of learned multi-scale intra-subject features for pretraining and develops several feature-wise regularizations that avoid collapsed identity representations; (2) During finetuning, it proposes a surprisingly simple self-supervised segmentation consistency regularization to exploit intra-subject correlation. Benchmarked in the one-shot segmentation setting, the proposed framework outperforms both well-tuned randomly-initialized baselines and current self-supervised techniques designed for both i.i.d. and longitudinal datasets. These improvements are demonstrated across both longitudinal neurodegenerative adult MRI and developing infant brain MRI and yield both higher performance and longitudinal consistency.
Improved Counting and Localization from Density Maps for Object Detection in 2D and 3D Microscopy Imaging
Mar 29, 2022



Abstract:Object counting and localization are key steps for quantitative analysis in large-scale microscopy applications. This procedure becomes challenging when target objects are overlapping, are densely clustered, and/or present fuzzy boundaries. Previous methods producing density maps based on deep learning have reached a high level of accuracy for object counting by assuming that object counting is equivalent to the integration of the density map. However, this model fails when objects show significant overlap regarding accurate localization. We propose an alternative method to count and localize objects from the density map to overcome this limitation. Our procedure includes the following three key aspects: 1) Proposing a new counting method based on the statistical properties of the density map, 2) optimizing the counting results for those objects which are well-detected based on the proposed counting method, and 3) improving localization of poorly detected objects using the proposed counting method as prior information. Validation includes processing of microscopy data with known ground truth and comparison with other models that use conventional processing of the density map. Our results show improved performance in counting and localization of objects in 2D and 3D microscopy data. Furthermore, the proposed method is generic, considering various applications that rely on the density map approach. Our code will be released post-review.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge