Axel Elaldi
Flash Invariant Point Attention
May 16, 2025



Abstract:Invariant Point Attention (IPA) is a key algorithm for geometry-aware modeling in structural biology, central to many protein and RNA models. However, its quadratic complexity limits the input sequence length. We introduce FlashIPA, a factorized reformulation of IPA that leverages hardware-efficient FlashAttention to achieve linear scaling in GPU memory and wall-clock time with sequence length. FlashIPA matches or exceeds standard IPA performance while substantially reducing computational costs. FlashIPA extends training to previously unattainable lengths, and we demonstrate this by re-training generative models without length restrictions and generating structures of thousands of residues. FlashIPA is available at https://github.com/flagshippioneering/flash_ipa.
Equivariant spatio-hemispherical networks for diffusion MRI deconvolution
Nov 18, 2024



Abstract:Each voxel in a diffusion MRI (dMRI) image contains a spherical signal corresponding to the direction and strength of water diffusion in the brain. This paper advances the analysis of such spatio-spherical data by developing convolutional network layers that are equivariant to the $\mathbf{E(3) \times SO(3)}$ group and account for the physical symmetries of dMRI including rotations, translations, and reflections of space alongside voxel-wise rotations. Further, neuronal fibers are typically antipodally symmetric, a fact we leverage to construct highly efficient spatio-hemispherical graph convolutions to accelerate the analysis of high-dimensional dMRI data. In the context of sparse spherical fiber deconvolution to recover white matter microstructure, our proposed equivariant network layers yield substantial performance and efficiency gains, leading to better and more practical resolution of crossing neuronal fibers and fiber tractography. These gains are experimentally consistent across both simulation and in vivo human datasets.
Bio2Token: All-atom tokenization of any biomolecular structure with Mamba
Oct 24, 2024



Abstract:Efficient encoding and representation of large 3D molecular structures with high fidelity is critical for biomolecular design applications. Despite this, many representation learning approaches restrict themselves to modeling smaller systems or use coarse-grained approximations of the systems, for example modeling proteins at the resolution of amino acid residues rather than at the level of individual atoms. To address this, we develop quantized auto-encoders that learn atom-level tokenizations of complete proteins, RNA and small molecule structures with reconstruction accuracies below and around 1 Angstrom. We demonstrate that the Mamba state space model architecture employed is comparatively efficient, requiring a fraction of the training data, parameters and compute needed to reach competitive accuracies and can scale to systems with almost 100,000 atoms. The learned structure tokens of bio2token may serve as the input for all-atom language models in the future.
$E \times SO$-Equivariant Networks for Spherical Deconvolution in Diffusion MRI
Apr 12, 2023Abstract:We present Roto-Translation Equivariant Spherical Deconvolution (RT-ESD), an $E(3)\times SO(3)$ equivariant framework for sparse deconvolution of volumes where each voxel contains a spherical signal. Such 6D data naturally arises in diffusion MRI (dMRI), a medical imaging modality widely used to measure microstructure and structural connectivity. As each dMRI voxel is typically a mixture of various overlapping structures, there is a need for blind deconvolution to recover crossing anatomical structures such as white matter tracts. Existing dMRI work takes either an iterative or deep learning approach to sparse spherical deconvolution, yet it typically does not account for relationships between neighboring measurements. This work constructs equivariant deep learning layers which respect to symmetries of spatial rotations, reflections, and translations, alongside the symmetries of voxelwise spherical rotations. As a result, RT-ESD improves on previous work across several tasks including fiber recovery on the DiSCo dataset, deconvolution-derived partial volume estimation on real-world \textit{in vivo} human brain dMRI, and improved downstream reconstruction of fiber tractograms on the Tractometer dataset. Our implementation is available at https://github.com/AxelElaldi/e3so3_conv
Equivariant Spherical Deconvolution: Learning Sparse Orientation Distribution Functions from Spherical Data
Feb 17, 2021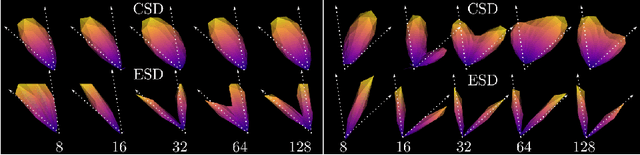
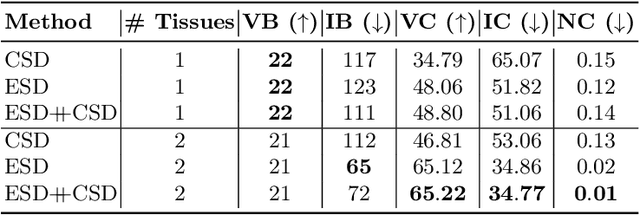
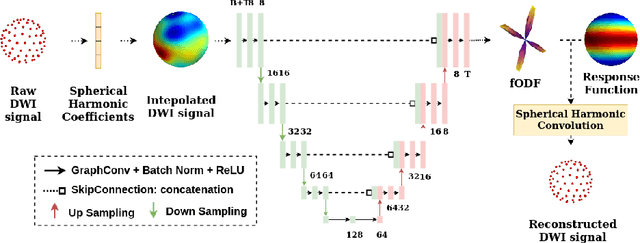
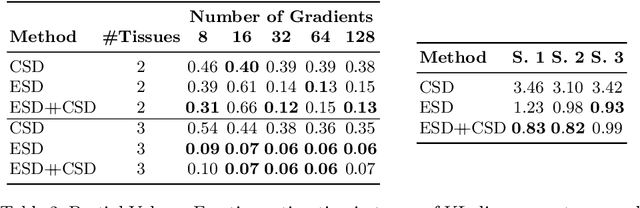
Abstract:We present a rotation-equivariant unsupervised learning framework for the sparse deconvolution of non-negative scalar fields defined on the unit sphere. Spherical signals with multiple peaks naturally arise in Diffusion MRI (dMRI), where each voxel consists of one or more signal sources corresponding to anisotropic tissue structure such as white matter. Due to spatial and spectral partial voluming, clinically-feasible dMRI struggles to resolve crossing-fiber white matter configurations, leading to extensive development in spherical deconvolution methodology to recover underlying fiber directions. However, these methods are typically linear and struggle with small crossing-angles and partial volume fraction estimation. In this work, we improve on current methodologies by nonlinearly estimating fiber structures via unsupervised spherical convolutional networks with guaranteed equivariance to spherical rotation. Experimentally, we first validate our proposition via extensive single and multi-shell synthetic benchmarks demonstrating competitive performance against common baselines. We then show improved downstream performance on fiber tractography measures on the Tractometer benchmark dataset. Finally, we show downstream improvements in terms of tractography and partial volume estimation on a multi-shell dataset of human subjects.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge