Andrew Liu
Tony
OpenAI GPT-5 System Card
Dec 19, 2025Abstract:This is the system card published alongside the OpenAI GPT-5 launch, August 2025. GPT-5 is a unified system with a smart and fast model that answers most questions, a deeper reasoning model for harder problems, and a real-time router that quickly decides which model to use based on conversation type, complexity, tool needs, and explicit intent (for example, if you say 'think hard about this' in the prompt). The router is continuously trained on real signals, including when users switch models, preference rates for responses, and measured correctness, improving over time. Once usage limits are reached, a mini version of each model handles remaining queries. This system card focuses primarily on gpt-5-thinking and gpt-5-main, while evaluations for other models are available in the appendix. The GPT-5 system not only outperforms previous models on benchmarks and answers questions more quickly, but -- more importantly -- is more useful for real-world queries. We've made significant advances in reducing hallucinations, improving instruction following, and minimizing sycophancy, and have leveled up GPT-5's performance in three of ChatGPT's most common uses: writing, coding, and health. All of the GPT-5 models additionally feature safe-completions, our latest approach to safety training to prevent disallowed content. Similarly to ChatGPT agent, we have decided to treat gpt-5-thinking as High capability in the Biological and Chemical domain under our Preparedness Framework, activating the associated safeguards. While we do not have definitive evidence that this model could meaningfully help a novice to create severe biological harm -- our defined threshold for High capability -- we have chosen to take a precautionary approach.
3D Roadway Scene Object Detection with LIDARs in Snowfall Conditions
Oct 25, 2025



Abstract:Because 3D structure of a roadway environment can be characterized directly by a Light Detection and Ranging (LiDAR) sensors, they can be used to obtain exceptional situational awareness for assitive and autonomous driving systems. Although LiDARs demonstrate good performance in clean and clear weather conditions, their performance significantly deteriorates in adverse weather conditions such as those involving atmospheric precipitation. This may render perception capabilities of autonomous systems that use LiDAR data in learning based models to perform object detection and ranging ineffective. While efforts have been made to enhance the accuracy of these models, the extent of signal degradation under various weather conditions remains largely not quantified. In this study, we focus on the performance of an automotive grade LiDAR in snowy conditions in order to develop a physics-based model that examines failure modes of a LiDAR sensor. Specifically, we investigated how the LiDAR signal attenuates with different snowfall rates and how snow particles near the source serve as small but efficient reflectors. Utilizing our model, we transform data from clear conditions to simulate snowy scenarios, enabling a comparison of our synthetic data with actual snowy conditions. Furthermore, we employ this synthetic data, representative of different snowfall rates, to explore the impact on a pre-trained object detection model, assessing its performance under varying levels of snowfall
Flash Invariant Point Attention
May 16, 2025



Abstract:Invariant Point Attention (IPA) is a key algorithm for geometry-aware modeling in structural biology, central to many protein and RNA models. However, its quadratic complexity limits the input sequence length. We introduce FlashIPA, a factorized reformulation of IPA that leverages hardware-efficient FlashAttention to achieve linear scaling in GPU memory and wall-clock time with sequence length. FlashIPA matches or exceeds standard IPA performance while substantially reducing computational costs. FlashIPA extends training to previously unattainable lengths, and we demonstrate this by re-training generative models without length restrictions and generating structures of thousands of residues. FlashIPA is available at https://github.com/flagshippioneering/flash_ipa.
FedFetch: Faster Federated Learning with Adaptive Downstream Prefetching
Apr 21, 2025Abstract:Federated learning (FL) is a machine learning paradigm that facilitates massively distributed model training with end-user data on edge devices directed by a central server. However, the large number of heterogeneous clients in FL deployments leads to a communication bottleneck between the server and the clients. This bottleneck is made worse by straggling clients, any one of which will further slow down training. To tackle these challenges, researchers have proposed techniques like client sampling and update compression. These techniques work well in isolation but combine poorly in the downstream, server-to-client direction. This is because unselected clients have outdated local model states and need to synchronize these states with the server first. We introduce FedFetch, a strategy to mitigate the download time overhead caused by combining client sampling and compression techniques. FedFetch achieves this with an efficient prefetch schedule for clients to prefetch model states multiple rounds before a stated training round. We empirically show that adding FedFetch to communication efficient FL techniques reduces end-to-end training time by 1.26$\times$ and download time by 4.49$\times$ across compression techniques with heterogeneous client settings. Our implementation is available at https://github.com/DistributedML/FedFetch
Bio2Token: All-atom tokenization of any biomolecular structure with Mamba
Oct 24, 2024



Abstract:Efficient encoding and representation of large 3D molecular structures with high fidelity is critical for biomolecular design applications. Despite this, many representation learning approaches restrict themselves to modeling smaller systems or use coarse-grained approximations of the systems, for example modeling proteins at the resolution of amino acid residues rather than at the level of individual atoms. To address this, we develop quantized auto-encoders that learn atom-level tokenizations of complete proteins, RNA and small molecule structures with reconstruction accuracies below and around 1 Angstrom. We demonstrate that the Mamba state space model architecture employed is comparatively efficient, requiring a fraction of the training data, parameters and compute needed to reach competitive accuracies and can scale to systems with almost 100,000 atoms. The learned structure tokens of bio2token may serve as the input for all-atom language models in the future.
A Role of Environmental Complexity on Representation Learning in Deep Reinforcement Learning Agents
Jul 03, 2024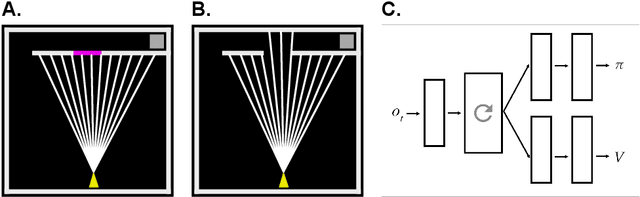
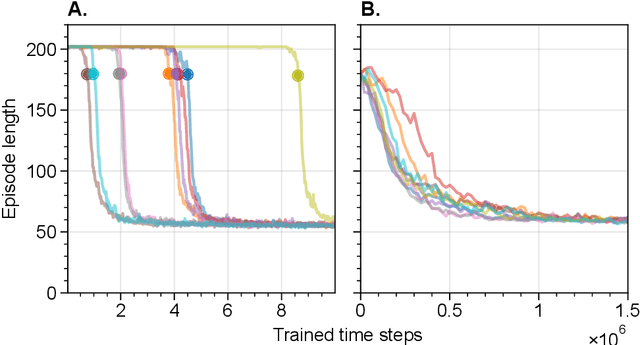
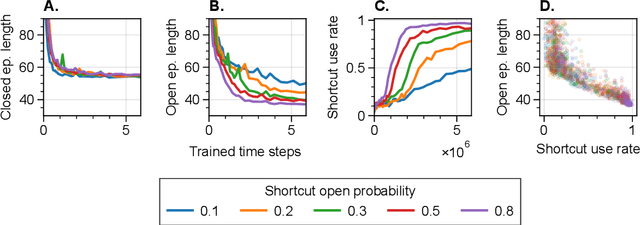
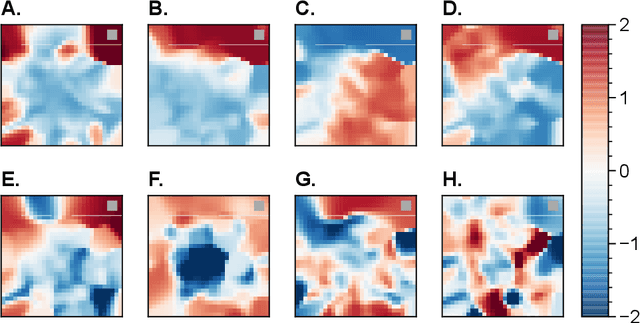
Abstract:The environments where individuals live can present diverse navigation challenges, resulting in varying navigation abilities and strategies. Inspired by differing urban layouts and the Dual Solutions Paradigm test used for human navigators, we developed a simulated navigation environment to train deep reinforcement learning agents in a shortcut usage task. We modulated the frequency of exposure to a shortcut and navigation cue, leading to the development of artificial agents with differing abilities. We examined the encoded representations in artificial neural networks driving these agents, revealing intricate dynamics in representation learning, and correlated them with shortcut use preferences. Furthermore, we demonstrated methods to analyze representations across a population of nodes, which proved effective in finding patterns in what would otherwise be noisy single-node data. These techniques may also have broader applications in studying neural activity. From our observations in representation learning dynamics, we propose insights for human navigation learning, emphasizing the importance of navigation challenges in developing strong landmark knowledge over repeated exposures to landmarks alone.
What does the Knowledge Neuron Thesis Have to do with Knowledge?
May 03, 2024



Abstract:We reassess the Knowledge Neuron (KN) Thesis: an interpretation of the mechanism underlying the ability of large language models to recall facts from a training corpus. This nascent thesis proposes that facts are recalled from the training corpus through the MLP weights in a manner resembling key-value memory, implying in effect that "knowledge" is stored in the network. Furthermore, by modifying the MLP modules, one can control the language model's generation of factual information. The plausibility of the KN thesis has been demonstrated by the success of KN-inspired model editing methods (Dai et al., 2022; Meng et al., 2022). We find that this thesis is, at best, an oversimplification. Not only have we found that we can edit the expression of certain linguistic phenomena using the same model editing methods but, through a more comprehensive evaluation, we have found that the KN thesis does not adequately explain the process of factual expression. While it is possible to argue that the MLP weights store complex patterns that are interpretable both syntactically and semantically, these patterns do not constitute "knowledge." To gain a more comprehensive understanding of the knowledge representation process, we must look beyond the MLP weights and explore recent models' complex layer structures and attention mechanisms.
Large Language Models in Healthcare: A Comprehensive Benchmark
Apr 25, 2024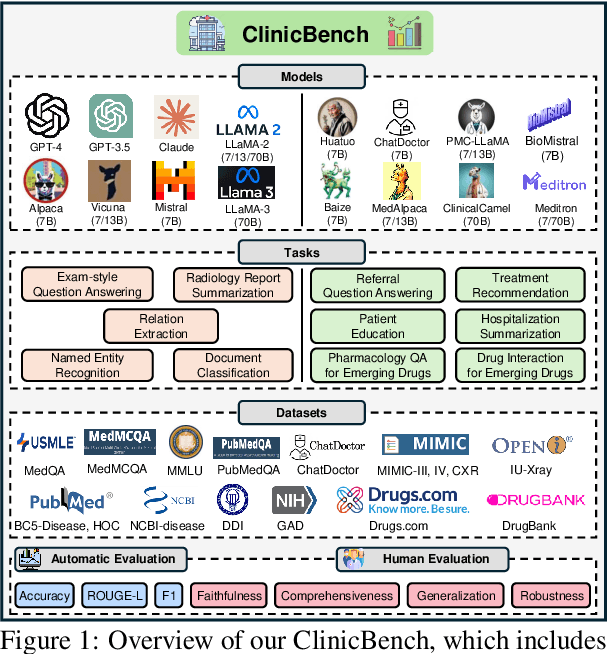



Abstract:The adoption of large language models (LLMs) to assist clinicians has attracted remarkable attention. Existing works mainly adopt the close-ended question-answering task with answer options for evaluation. However, in real clinical settings, many clinical decisions, such as treatment recommendations, involve answering open-ended questions without pre-set options. Meanwhile, existing studies mainly use accuracy to assess model performance. In this paper, we comprehensively benchmark diverse LLMs in healthcare, to clearly understand their strengths and weaknesses. Our benchmark contains seven tasks and thirteen datasets across medical language generation, understanding, and reasoning. We conduct a detailed evaluation of the existing sixteen LLMs in healthcare under both zero-shot and few-shot (i.e., 1,3,5-shot) learning settings. We report the results on five metrics (i.e. matching, faithfulness, comprehensiveness, generalizability, and robustness) that are critical in achieving trust from clinical users. We further invite medical experts to conduct human evaluation.
Qilin-Med: Multi-stage Knowledge Injection Advanced Medical Large Language Model
Oct 13, 2023



Abstract:Integrating large language models (LLMs) into healthcare presents potential but faces challenges. Directly pre-training LLMs for domains like medicine is resource-heavy and sometimes unfeasible. Sole reliance on Supervised Fine-tuning (SFT) can result in overconfident predictions and may not tap into domain specific insights. Addressing these challenges, we present a multi-stage training method combining Domain-specific Continued Pre-training (DCPT), SFT, and Direct Preference Optimization (DPO). A notable contribution of our study is the introduction of a 3Gb Chinese Medicine (ChiMed) dataset, encompassing medical question answering, plain texts, knowledge graphs, and dialogues, segmented into three training stages. The medical LLM trained with our pipeline, Qilin-Med, exhibits significant performance boosts. In the CPT and SFT phases, it achieves 38.4% and 40.0% accuracy on the CMExam, surpassing Baichuan-7B's 33.5%. In the DPO phase, on the Huatuo-26M test set, it scores 16.66 in BLEU-1 and 27.44 in ROUGE1, outperforming the SFT's 12.69 and 24.21. This highlights the strength of our training approach in refining LLMs for medical applications.
Benchmarking Large Language Models on CMExam -- A Comprehensive Chinese Medical Exam Dataset
Jun 08, 2023



Abstract:Recent advancements in large language models (LLMs) have transformed the field of question answering (QA). However, evaluating LLMs in the medical field is challenging due to the lack of standardized and comprehensive datasets. To address this gap, we introduce CMExam, sourced from the Chinese National Medical Licensing Examination. CMExam consists of 60K+ multiple-choice questions for standardized and objective evaluations, as well as solution explanations for model reasoning evaluation in an open-ended manner. For in-depth analyses of LLMs, we invited medical professionals to label five additional question-wise annotations, including disease groups, clinical departments, medical disciplines, areas of competency, and question difficulty levels. Alongside the dataset, we further conducted thorough experiments with representative LLMs and QA algorithms on CMExam. The results show that GPT-4 had the best accuracy of 61.6% and a weighted F1 score of 0.617. These results highlight a great disparity when compared to human accuracy, which stood at 71.6%. For explanation tasks, while LLMs could generate relevant reasoning and demonstrate improved performance after finetuning, they fall short of a desired standard, indicating ample room for improvement. To the best of our knowledge, CMExam is the first Chinese medical exam dataset to provide comprehensive medical annotations. The experiments and findings of LLM evaluation also provide valuable insights into the challenges and potential solutions in developing Chinese medical QA systems and LLM evaluation pipelines. The dataset and relevant code are available at https://github.com/williamliujl/CMExam.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge