Jo Schlemper
Dual-Domain Self-Supervised Learning for Accelerated Non-Cartesian MRI Reconstruction
Feb 18, 2023



Abstract:While enabling accelerated acquisition and improved reconstruction accuracy, current deep MRI reconstruction networks are typically supervised, require fully sampled data, and are limited to Cartesian sampling patterns. These factors limit their practical adoption as fully-sampled MRI is prohibitively time-consuming to acquire clinically. Further, non-Cartesian sampling patterns are particularly desirable as they are more amenable to acceleration and show improved motion robustness. To this end, we present a fully self-supervised approach for accelerated non-Cartesian MRI reconstruction which leverages self-supervision in both k-space and image domains. In training, the undersampled data are split into disjoint k-space domain partitions. For the k-space self-supervision, we train a network to reconstruct the input undersampled data from both the disjoint partitions and from itself. For the image-level self-supervision, we enforce appearance consistency obtained from the original undersampled data and the two partitions. Experimental results on our simulated multi-coil non-Cartesian MRI dataset demonstrate that DDSS can generate high-quality reconstruction that approaches the accuracy of the fully supervised reconstruction, outperforming previous baseline methods. Finally, DDSS is shown to scale to highly challenging real-world clinical MRI reconstruction acquired on a portable low-field (0.064 T) MRI scanner with no data available for supervised training while demonstrating improved image quality as compared to traditional reconstruction, as determined by a radiologist study.
ContraReg: Contrastive Learning of Multi-modality Unsupervised Deformable Image Registration
Jun 27, 2022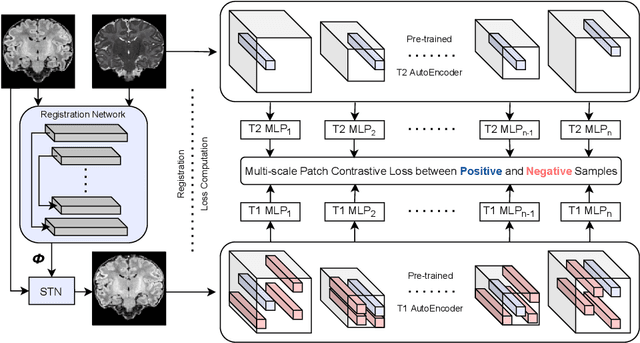
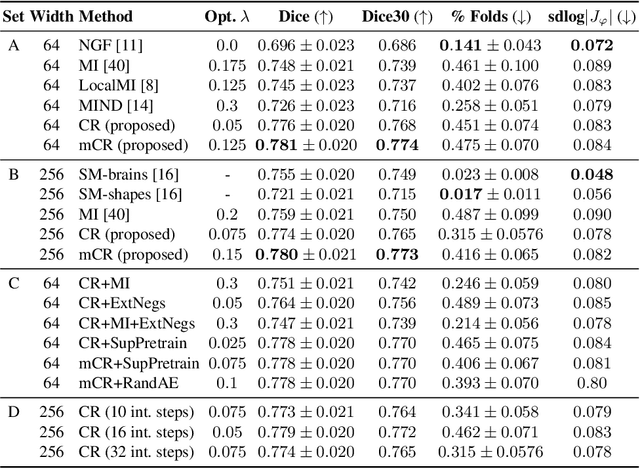
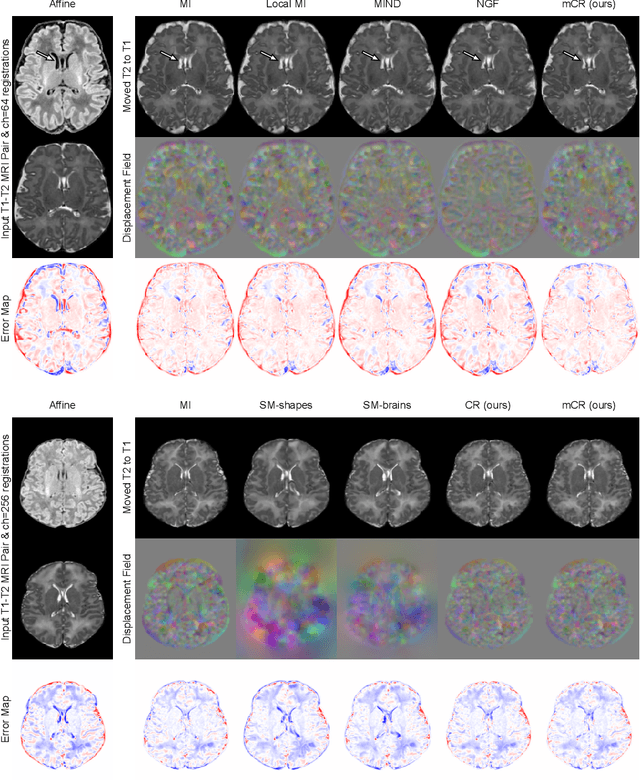
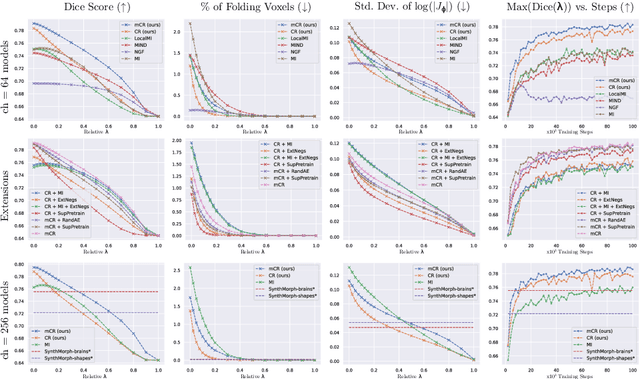
Abstract:Establishing voxelwise semantic correspondence across distinct imaging modalities is a foundational yet formidable computer vision task. Current multi-modality registration techniques maximize hand-crafted inter-domain similarity functions, are limited in modeling nonlinear intensity-relationships and deformations, and may require significant re-engineering or underperform on new tasks, datasets, and domain pairs. This work presents ContraReg, an unsupervised contrastive representation learning approach to multi-modality deformable registration. By projecting learned multi-scale local patch features onto a jointly learned inter-domain embedding space, ContraReg obtains representations useful for non-rigid multi-modality alignment. Experimentally, ContraReg achieves accurate and robust results with smooth and invertible deformations across a series of baselines and ablations on a neonatal T1-T2 brain MRI registration task with all methods validated over a wide range of deformation regularization strengths.
DSFormer: A Dual-domain Self-supervised Transformer for Accelerated Multi-contrast MRI Reconstruction
Jan 26, 2022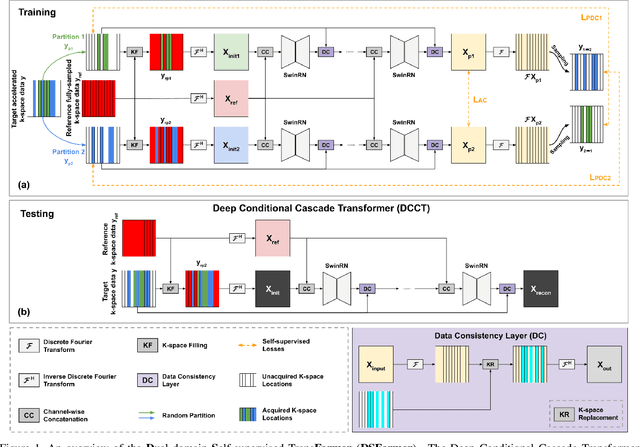
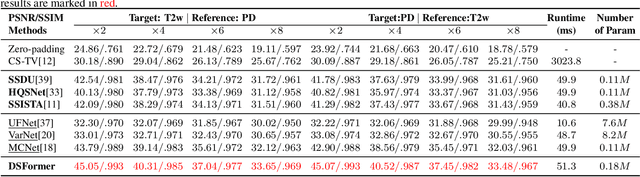
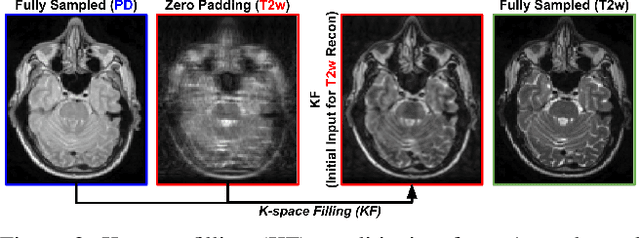
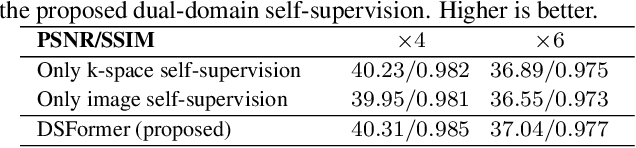
Abstract:Multi-contrast MRI (MC-MRI) captures multiple complementary imaging modalities to aid in radiological decision-making. Given the need for lowering the time cost of multiple acquisitions, current deep accelerated MRI reconstruction networks focus on exploiting the redundancy between multiple contrasts. However, existing works are largely supervised with paired data and/or prohibitively expensive fully-sampled MRI sequences. Further, reconstruction networks typically rely on convolutional architectures which are limited in their capacity to model long-range interactions and may lead to suboptimal recovery of fine anatomical detail. To these ends, we present a dual-domain self-supervised transformer (DSFormer) for accelerated MC-MRI reconstruction. DSFormer develops a deep conditional cascade transformer (DCCT) consisting of several cascaded Swin transformer reconstruction networks (SwinRN) trained under two deep conditioning strategies to enable MC-MRI information sharing. We further present a dual-domain (image and k-space) self-supervised learning strategy for DCCT to alleviate the costs of acquiring fully sampled training data. DSFormer generates high-fidelity reconstructions which experimentally outperform current fully-supervised baselines. Moreover, we find that DSFormer achieves nearly the same performance when trained either with full supervision or with our proposed dual-domain self-supervision.
Complementary Time-Frequency Domain Networks for Dynamic Parallel MR Image Reconstruction
Dec 22, 2020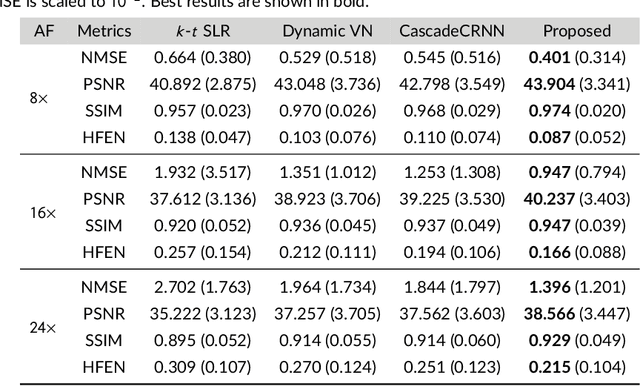
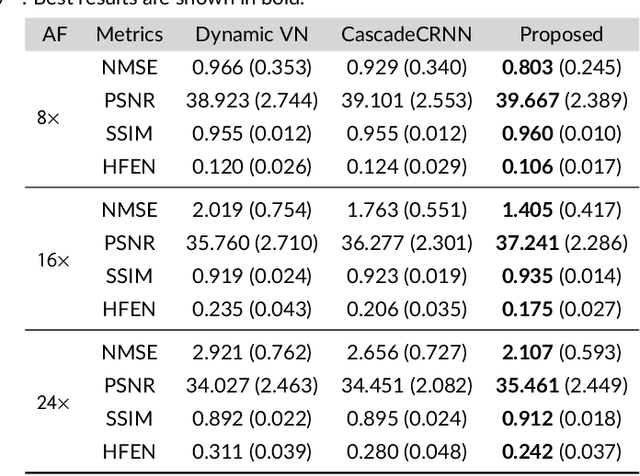
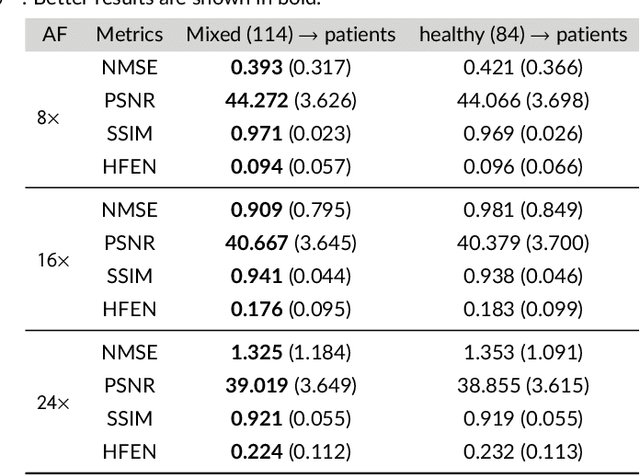
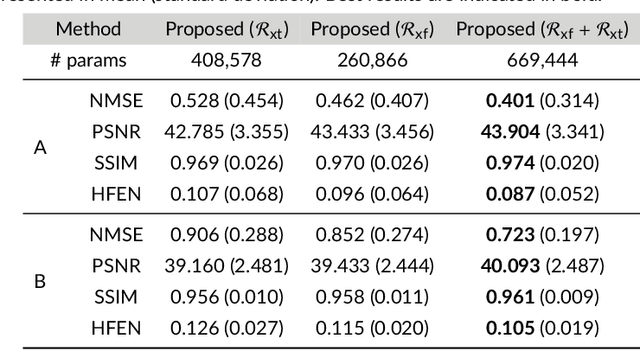
Abstract:Purpose: To introduce a novel deep learning based approach for fast and high-quality dynamic multi-coil MR reconstruction by learning a complementary time-frequency domain network that exploits spatio-temporal correlations simultaneously from complementary domains. Theory and Methods: Dynamic parallel MR image reconstruction is formulated as a multi-variable minimisation problem, where the data is regularised in combined temporal Fourier and spatial (x-f) domain as well as in spatio-temporal image (x-t) domain. An iterative algorithm based on variable splitting technique is derived, which alternates among signal de-aliasing steps in x-f and x-t spaces, a closed-form point-wise data consistency step and a weighted coupling step. The iterative model is embedded into a deep recurrent neural network which learns to recover the image via exploiting spatio-temporal redundancies in complementary domains. Results: Experiments were performed on two datasets of highly undersampled multi-coil short-axis cardiac cine MRI scans. Results demonstrate that our proposed method outperforms the current state-of-the-art approaches both quantitatively and qualitatively. The proposed model can also generalise well to data acquired from a different scanner and data with pathologies that were not seen in the training set. Conclusion: The work shows the benefit of reconstructing dynamic parallel MRI in complementary time-frequency domains with deep neural networks. The method can effectively and robustly reconstruct high-quality images from highly undersampled dynamic multi-coil data ($16 \times$ and $24 \times$ yielding 15s and 10s scan times respectively) with fast reconstruction speed (2.8s). This could potentially facilitate achieving fast single-breath-hold clinical 2D cardiac cine imaging.
Deep Network Interpolation for Accelerated Parallel MR Image Reconstruction
Jul 12, 2020


Abstract:We present a deep network interpolation strategy for accelerated parallel MR image reconstruction. In particular, we examine the network interpolation in parameter space between a source model that is formulated in an unrolled scheme with L1 and SSIM losses and its counterpart that is trained with an adversarial loss. We show that by interpolating between the two different models of the same network structure, the new interpolated network can model a trade-off between perceptual quality and fidelity.
$Σ$-net: Systematic Evaluation of Iterative Deep Neural Networks for Fast Parallel MR Image Reconstruction
Dec 18, 2019



Abstract:Purpose: To systematically investigate the influence of various data consistency layers, (semi-)supervised learning and ensembling strategies, defined in a $\Sigma$-net, for accelerated parallel MR image reconstruction using deep learning. Theory and Methods: MR image reconstruction is formulated as learned unrolled optimization scheme with a Down-Up network as regularization and varying data consistency layers. The different architectures are split into sensitivity networks, which rely on explicit coil sensitivity maps, and parallel coil networks, which learn the combination of coils implicitly. Different content and adversarial losses, a semi-supervised fine-tuning scheme and model ensembling are investigated. Results: Evaluated on the fastMRI multicoil validation set, architectures involving raw k-space data outperform image enhancement methods significantly. Semi-supervised fine-tuning adapts to new k-space data and provides, together with reconstructions based on adversarial training, the visually most appealing results although quantitative quality metrics are reduced. The $\Sigma$-net ensembles the benefits from different models and achieves similar scores compared to the single state-of-the-art approaches. Conclusion: This work provides an open-source framework to perform a systematic wide-range comparison of state-of-the-art reconstruction approaches for parallel MR image reconstruction on the fastMRI knee dataset and explores the importance of data consistency. A suitable trade-off between perceptual image quality and quantitative scores are achieved with the ensembled $\Sigma$-net.
$Σ$-net: Ensembled Iterative Deep Neural Networks for Accelerated Parallel MR Image Reconstruction
Dec 11, 2019

Abstract:We explore an ensembled $\Sigma$-net for fast parallel MR imaging, including parallel coil networks, which perform implicit coil weighting, and sensitivity networks, involving explicit sensitivity maps. The networks in $\Sigma$-net are trained in a supervised way, including content and GAN losses, and with various ways of data consistency, i.e., proximal mappings, gradient descent and variable splitting. A semi-supervised finetuning scheme allows us to adapt to the k-space data at test time, which, however, decreases the quantitative metrics, although generating the visually most textured and sharp images. For this challenge, we focused on robust and high SSIM scores, which we achieved by ensembling all models to a $\Sigma$-net.
Data consistency networks for (calibration-less) accelerated parallel MR image reconstruction
Sep 25, 2019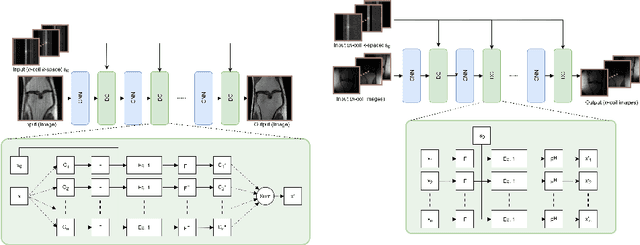
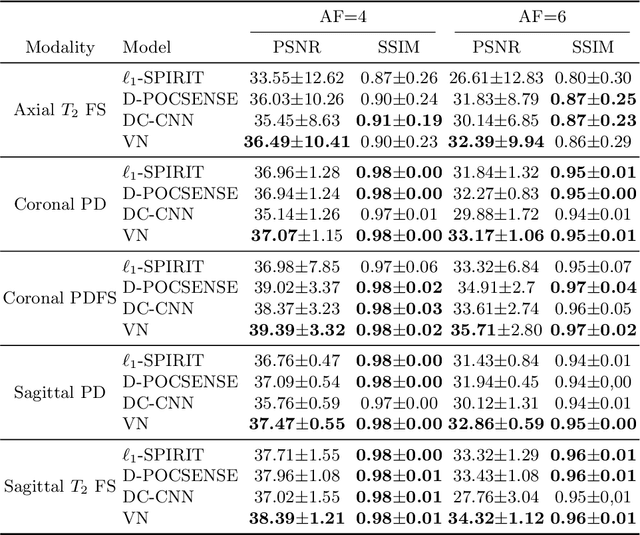
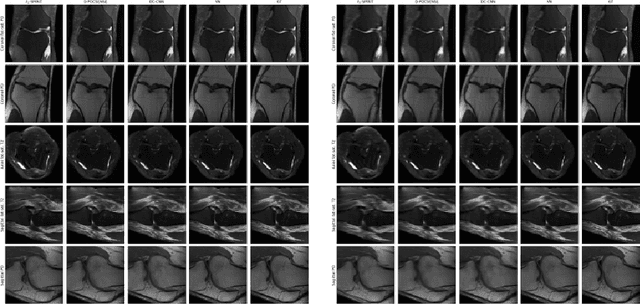
Abstract:We present simple reconstruction networks for multi-coil data by extending deep cascade of CNN's and exploiting the data consistency layer. In particular, we propose two variants, where one is inspired by POCSENSE and the other is calibration-less. We show that the proposed approaches are competitive relative to the state of the art both quantitatively and qualitatively.
dAUTOMAP: decomposing AUTOMAP to achieve scalability and enhance performance
Sep 25, 2019



Abstract:AUTOMAP is a promising generalized reconstruction approach, however, it is not scalable and hence the practicality is limited. We present dAUTOMAP, a novel way for decomposing the domain transformation of AUTOMAP, making the model scale linearly. We show dAUTOMAP outperforms AUTOMAP with significantly fewer parameters.
Unsupervised Multi-modal Style Transfer for Cardiac MR Segmentation
Aug 21, 2019



Abstract:In this work, we present a fully automatic method to segment cardiac structures from late-gadolinium enhanced (LGE) images without using labelled LGE data for training, but instead by transferring the anatomical knowledge and features learned on annotated balanced steady-state free precession (bSSFP) images, which are easier to acquire. Our framework mainly consists of two neural networks: a multi-modal image translation network for style transfer and a cascaded segmentation network for image segmentation. The multi-modal image translation network generates realistic and diverse synthetic LGE images conditioned on a single annotated bSSFP image, forming a synthetic LGE training set. This set is then utilized to fine-tune the segmentation network pre-trained on labelled bSSFP images, achieving the goal of unsupervised LGE image segmentation. In particular, the proposed cascaded segmentation network is able to produce accurate segmentation by taking both shape prior and image appearance into account, achieving an average Dice score of 0.92 for the left ventricle, 0.83 for the myocardium, and 0.88 for the right ventricle on the test set.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge