Thomas Ach
Microscopy Image Segmentation via Point and Shape Regularized Data Synthesis
Aug 18, 2023Abstract:Current deep learning-based approaches for the segmentation of microscopy images heavily rely on large amount of training data with dense annotation, which is highly costly and laborious in practice. Compared to full annotation where the complete contour of objects is depicted, point annotations, specifically object centroids, are much easier to acquire and still provide crucial information about the objects for subsequent segmentation. In this paper, we assume access to point annotations only during training and develop a unified pipeline for microscopy image segmentation using synthetically generated training data. Our framework includes three stages: (1) it takes point annotations and samples a pseudo dense segmentation mask constrained with shape priors; (2) with an image generative model trained in an unpaired manner, it translates the mask to a realistic microscopy image regularized by object level consistency; (3) the pseudo masks along with the synthetic images then constitute a pairwise dataset for training an ad-hoc segmentation model. On the public MoNuSeg dataset, our synthesis pipeline produces more diverse and realistic images than baseline models while maintaining high coherence between input masks and generated images. When using the identical segmentation backbones, the models trained on our synthetic dataset significantly outperform those trained with pseudo-labels or baseline-generated images. Moreover, our framework achieves comparable results to models trained on authentic microscopy images with dense labels, demonstrating its potential as a reliable and highly efficient alternative to labor-intensive manual pixel-wise annotations in microscopy image segmentation. The code is available.
Improved Counting and Localization from Density Maps for Object Detection in 2D and 3D Microscopy Imaging
Mar 29, 2022



Abstract:Object counting and localization are key steps for quantitative analysis in large-scale microscopy applications. This procedure becomes challenging when target objects are overlapping, are densely clustered, and/or present fuzzy boundaries. Previous methods producing density maps based on deep learning have reached a high level of accuracy for object counting by assuming that object counting is equivalent to the integration of the density map. However, this model fails when objects show significant overlap regarding accurate localization. We propose an alternative method to count and localize objects from the density map to overcome this limitation. Our procedure includes the following three key aspects: 1) Proposing a new counting method based on the statistical properties of the density map, 2) optimizing the counting results for those objects which are well-detected based on the proposed counting method, and 3) improving localization of poorly detected objects using the proposed counting method as prior information. Validation includes processing of microscopy data with known ground truth and comparison with other models that use conventional processing of the density map. Our results show improved performance in counting and localization of objects in 2D and 3D microscopy data. Furthermore, the proposed method is generic, considering various applications that rely on the density map approach. Our code will be released post-review.
Point-supervised Segmentation of Microscopy Images and Volumes via Objectness Regularization
Mar 19, 2021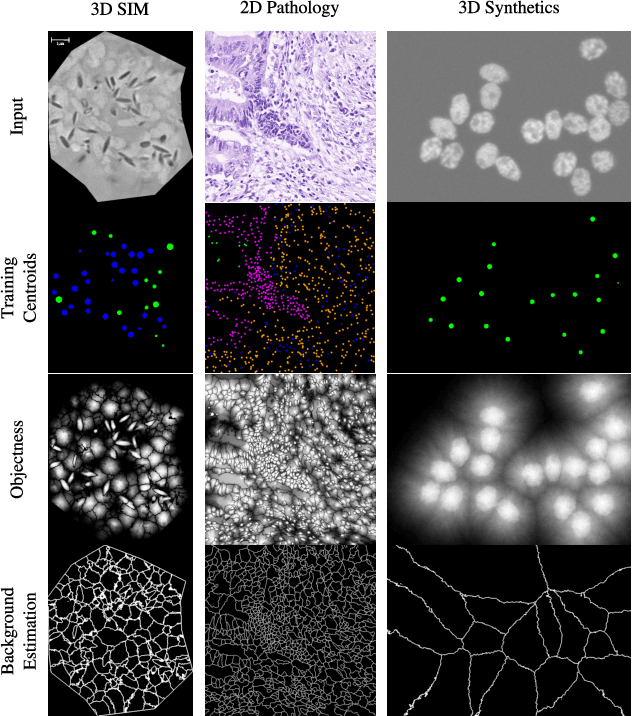
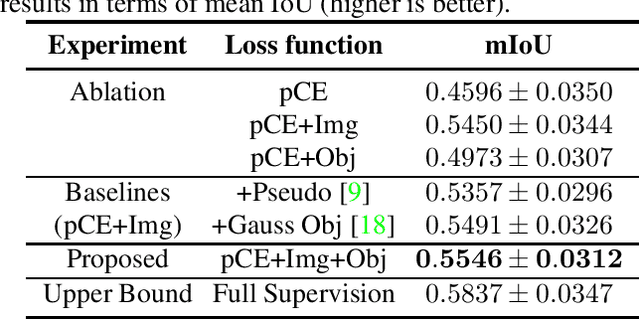
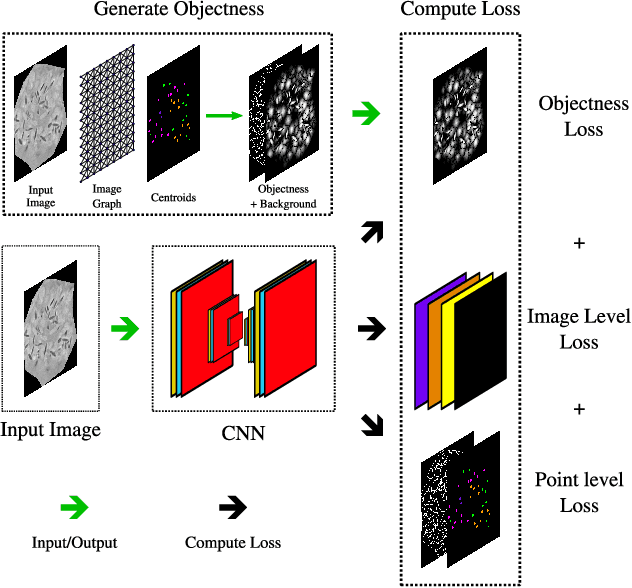
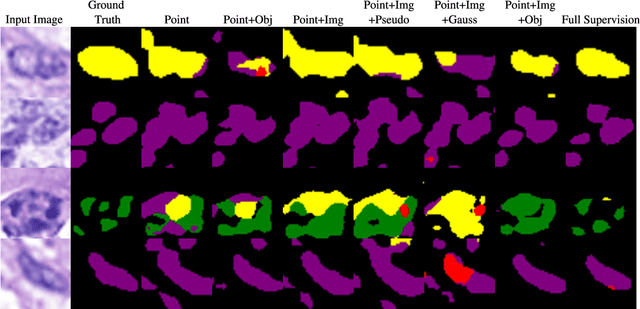
Abstract:Annotation is a major hurdle in the semantic segmentation of microscopy images and volumes due to its prerequisite expertise and effort. This work enables the training of semantic segmentation networks on images with only a single point for training per instance, an extreme case of weak supervision which drastically reduces the burden of annotation. Our approach has two key aspects: (1) we construct a graph-theoretic soft-segmentation using individual seeds to be used within a regularizer during training and (2) we use an objective function that enables learning from the constructed soft-labels. We achieve competitive results against the state-of-the-art in point-supervised semantic segmentation on challenging datasets in digital pathology. Finally, we scale our methodology to point-supervised segmentation in 3D fluorescence microscopy volumes, obviating the need for arduous manual volumetric delineation. Our code is freely available.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge