Leo Milecki
Effective Segmentation of Post-Treatment Gliomas Using Simple Approaches: Artificial Sequence Generation and Ensemble Models
Sep 12, 2024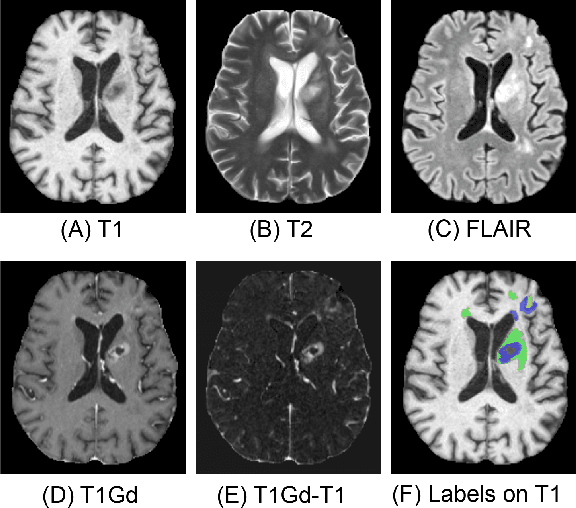
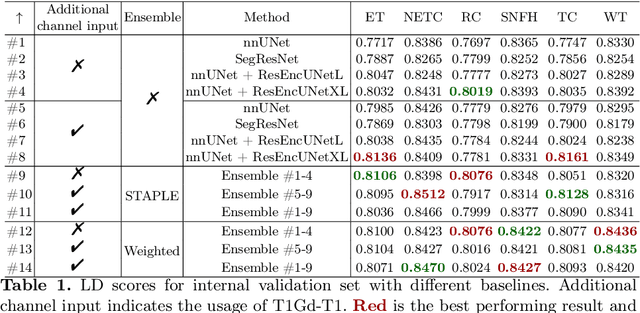
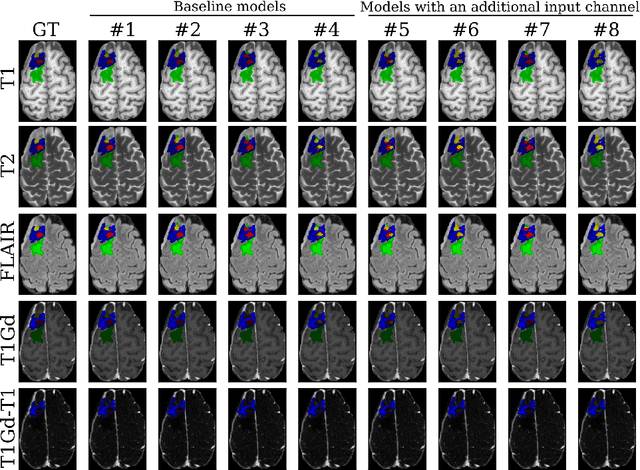
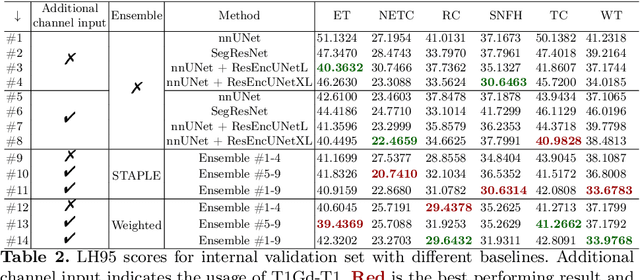
Abstract:Segmentation is a crucial task in the medical imaging field and is often an important primary step or even a prerequisite to the analysis of medical volumes. Yet treatments such as surgery complicate the accurate delineation of regions of interest. The BraTS Post-Treatment 2024 Challenge published the first public dataset for post-surgery glioma segmentation and addresses the aforementioned issue by fostering the development of automated segmentation tools for glioma in MRI data. In this effort, we propose two straightforward approaches to enhance the segmentation performances of deep learning-based methodologies. First, we incorporate an additional input based on a simple linear combination of the available MRI sequences input, which highlights enhancing tumors. Second, we employ various ensembling methods to weigh the contribution of a battery of models. Our results demonstrate that these approaches significantly improve segmentation performance compared to baseline models, underscoring the effectiveness of these simple approaches in improving medical image segmentation tasks.
ADAPT: Multimodal Learning for Detecting Physiological Changes under Missing Modalities
Jul 04, 2024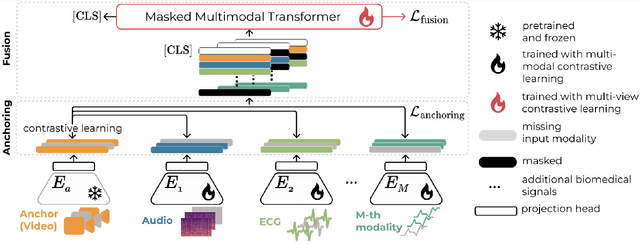
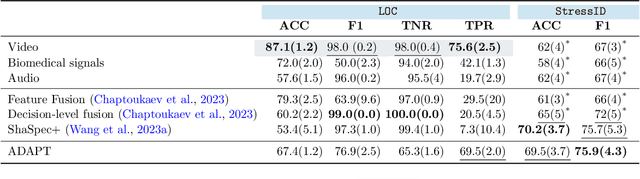
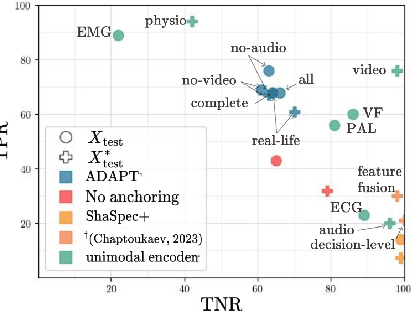
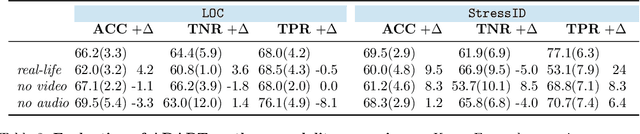
Abstract:Multimodality has recently gained attention in the medical domain, where imaging or video modalities may be integrated with biomedical signals or health records. Yet, two challenges remain: balancing the contributions of modalities, especially in cases with a limited amount of data available, and tackling missing modalities. To address both issues, in this paper, we introduce the AnchoreD multimodAl Physiological Transformer (ADAPT), a multimodal, scalable framework with two key components: (i) aligning all modalities in the space of the strongest, richest modality (called anchor) to learn a joint embedding space, and (ii) a Masked Multimodal Transformer, leveraging both inter- and intra-modality correlations while handling missing modalities. We focus on detecting physiological changes in two real-life scenarios: stress in individuals induced by specific triggers and fighter pilots' loss of consciousness induced by $g$-forces. We validate the generalizability of ADAPT through extensive experiments on two datasets for these tasks, where we set the new state of the art while demonstrating its robustness across various modality scenarios and its high potential for real-life applications.
MEDIMP: Medical Images and Prompts for renal transplant representation learning
Mar 22, 2023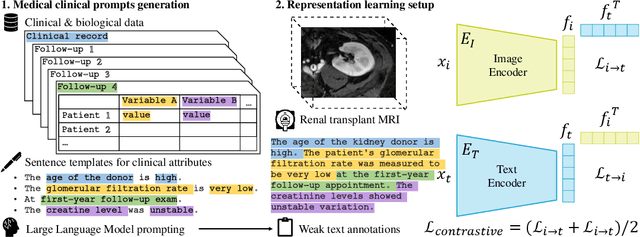
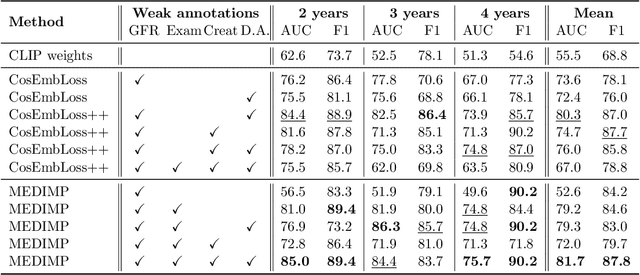
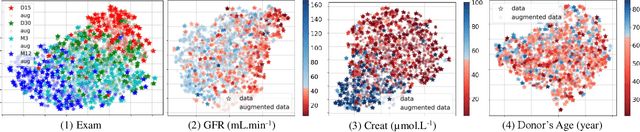
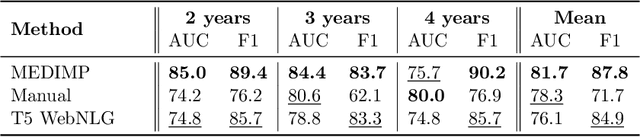
Abstract:Renal transplantation emerges as the most effective solution for end-stage renal disease. Occurring from complex causes, a substantial risk of transplant chronic dysfunction persists and may lead to graft loss. Medical imaging plays a substantial role in renal transplant monitoring in clinical practice. However, graft supervision is multi-disciplinary, notably joining nephrology, urology, and radiology, while identifying robust biomarkers from such high-dimensional and complex data for prognosis is challenging. In this work, taking inspiration from the recent success of Large Language Models (LLMs), we propose MEDIMP -- Medical Images and Prompts -- a model to learn meaningful multi-modal representations of renal transplant Dynamic Contrast-Enhanced Magnetic Resonance Imaging (DCE MRI) by incorporating structural clinicobiological data after translating them into text prompts. MEDIMP is based on contrastive learning from joint text-image paired embeddings to perform this challenging task. Moreover, we propose a framework that generates medical prompts using automatic textual data augmentations from LLMs. Our goal is to learn meaningful manifolds of renal transplant DCE MRI, interesting for the prognosis of the transplant or patient status (2, 3, and 4 years after the transplant), fully exploiting the available multi-modal data in the most efficient way. Extensive experiments and comparisons with other renal transplant representation learning methods with limited data prove the effectiveness of MEDIMP in a relevant clinical setting, giving new directions toward medical prompts. Our code is available at https://github.com/leomlck/MEDIMP.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge