Image segmentation of liver stage malaria infection with spatial uncertainty sampling
Paper and Code
Nov 30, 2019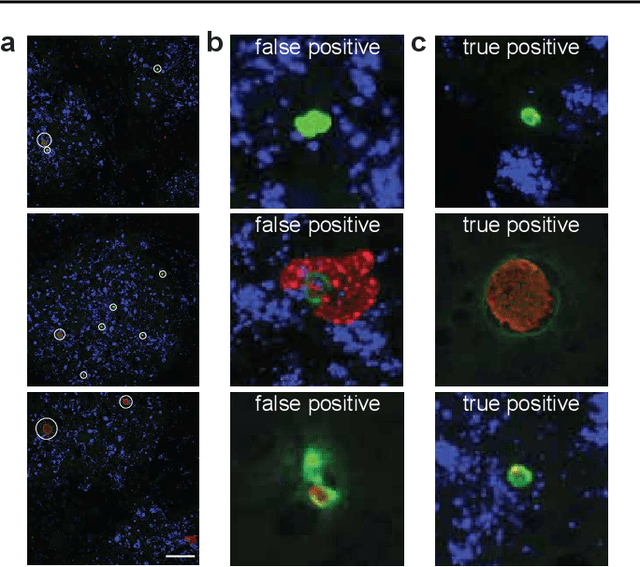
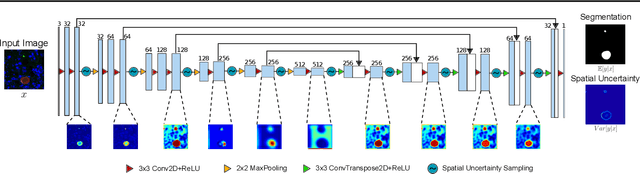
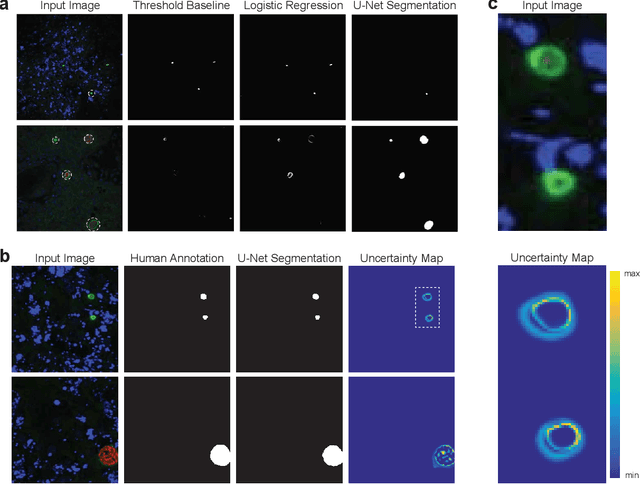
Global eradication of malaria depends on the development of drugs effective against the silent, yet obligate liver stage of the disease. The gold standard in drug development remains microscopic imaging of liver stage parasites in in vitro cell culture models. Image analysis presents a major bottleneck in this pipeline since the parasite has significant variability in size, shape, and density in these models. As with other highly variable datasets, traditional segmentation models have poor generalizability as they rely on hand-crafted features; thus, manual annotation of liver stage malaria images remains standard. To address this need, we develop a convolutional neural network architecture that utilizes spatial dropout sampling for parasite segmentation and epistemic uncertainty estimation in images of liver stage malaria. Our pipeline produces high-precision segmentations nearly identical to expert annotations, generalizes well on a diverse dataset of liver stage malaria parasites, and promotes independence between learned feature maps to model the uncertainty of generated predictions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge