Wolfgang Bogner
High-Field MR Center, Department of Biomedical Imaging and Image-guided Therapy, Medical University of Vienna, Vienna, Austria
A Flow-based Truncated Denoising Diffusion Model for Super-resolution Magnetic Resonance Spectroscopic Imaging
Oct 25, 2024Abstract:Magnetic Resonance Spectroscopic Imaging (MRSI) is a non-invasive imaging technique for studying metabolism and has become a crucial tool for understanding neurological diseases, cancers and diabetes. High spatial resolution MRSI is needed to characterize lesions, but in practice MRSI is acquired at low resolution due to time and sensitivity restrictions caused by the low metabolite concentrations. Therefore, there is an imperative need for a post-processing approach to generate high-resolution MRSI from low-resolution data that can be acquired fast and with high sensitivity. Deep learning-based super-resolution methods provided promising results for improving the spatial resolution of MRSI, but they still have limited capability to generate accurate and high-quality images. Recently, diffusion models have demonstrated superior learning capability than other generative models in various tasks, but sampling from diffusion models requires iterating through a large number of diffusion steps, which is time-consuming. This work introduces a Flow-based Truncated Denoising Diffusion Model (FTDDM) for super-resolution MRSI, which shortens the diffusion process by truncating the diffusion chain, and the truncated steps are estimated using a normalizing flow-based network. The network is conditioned on upscaling factors to enable multi-scale super-resolution. To train and evaluate the deep learning models, we developed a 1H-MRSI dataset acquired from 25 high-grade glioma patients. We demonstrate that FTDDM outperforms existing generative models while speeding up the sampling process by over 9-fold compared to the baseline diffusion model. Neuroradiologists' evaluations confirmed the clinical advantages of our method, which also supports uncertainty estimation and sharpness adjustment, extending its potential clinical applications.
* Accepted by Medical Image Analysis (MedIA)
WALINET: A water and lipid identification convolutional Neural Network for nuisance signal removal in 1H MR Spectroscopic Imaging
Oct 01, 2024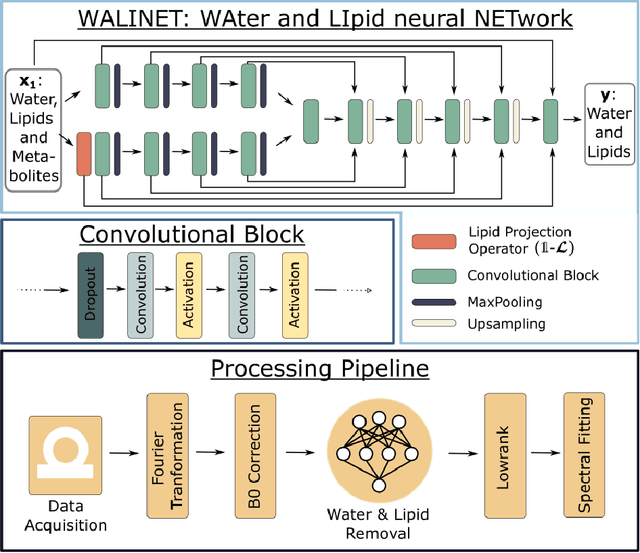

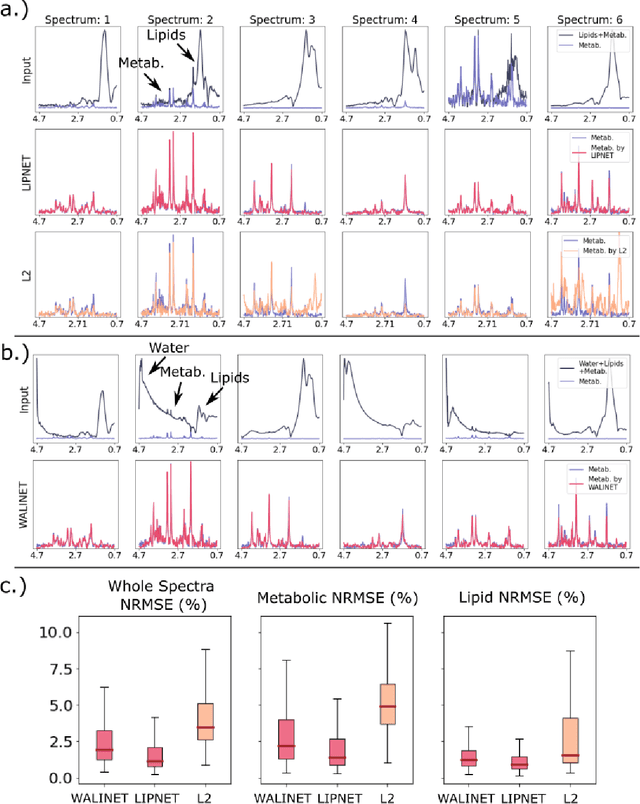
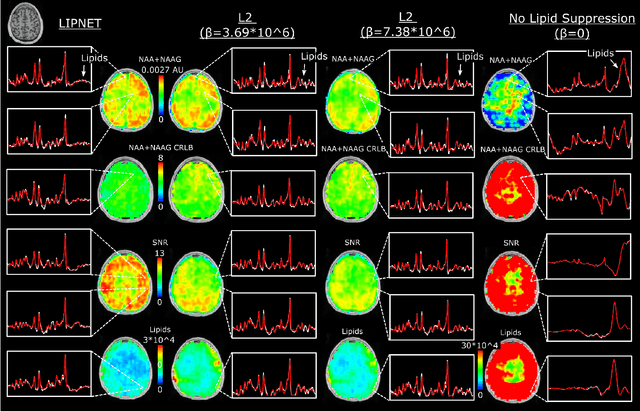
Abstract:Purpose. Proton Magnetic Resonance Spectroscopic Imaging (1H-MRSI) provides non-invasive spectral-spatial mapping of metabolism. However, long-standing problems in whole-brain 1H-MRSI are spectral overlap of metabolite peaks with large lipid signal from scalp, and overwhelming water signal that distorts spectra. Fast and effective methods are needed for high-resolution 1H-MRSI to accurately remove lipid and water signals while preserving the metabolite signal. The potential of supervised neural networks for this task remains unexplored, despite their success for other MRSI processing. Methods. We introduce a deep-learning method based on a modified Y-NET network for water and lipid removal in whole-brain 1H-MRSI. The WALINET (WAter and LIpid neural NETwork) was compared to conventional methods such as the state-of-the-art lipid L2 regularization and Hankel-Lanczos singular value decomposition (HLSVD) water suppression. Methods were evaluated on simulated and in-vivo whole-brain MRSI using NMRSE, SNR, CRLB, and FWHM metrics. Results. WALINET is significantly faster and needs 8s for high-resolution whole-brain MRSI, compared to 42 minutes for conventional HLSVD+L2. Quantitative analysis shows WALINET has better performance than HLSVD+L2: 1) more lipid removal with 41% lower NRMSE, 2) better metabolite signal preservation with 71% lower NRMSE in simulated data, 155% higher SNR and 50% lower CRLB in in-vivo data. Metabolic maps obtained by WALINET in healthy subjects and patients show better gray/white-matter contrast with more visible structural details. Conclusions. WALINET has superior performance for nuisance signal removal and metabolite quantification on whole-brain 1H-MRSI compared to conventional state-of-the-art techniques. This represents a new application of deep-learning for MRSI processing, with potential for automated high-throughput workflow.
Deep-ER: Deep Learning ECCENTRIC Reconstruction for fast high-resolution neurometabolic imaging
Sep 26, 2024
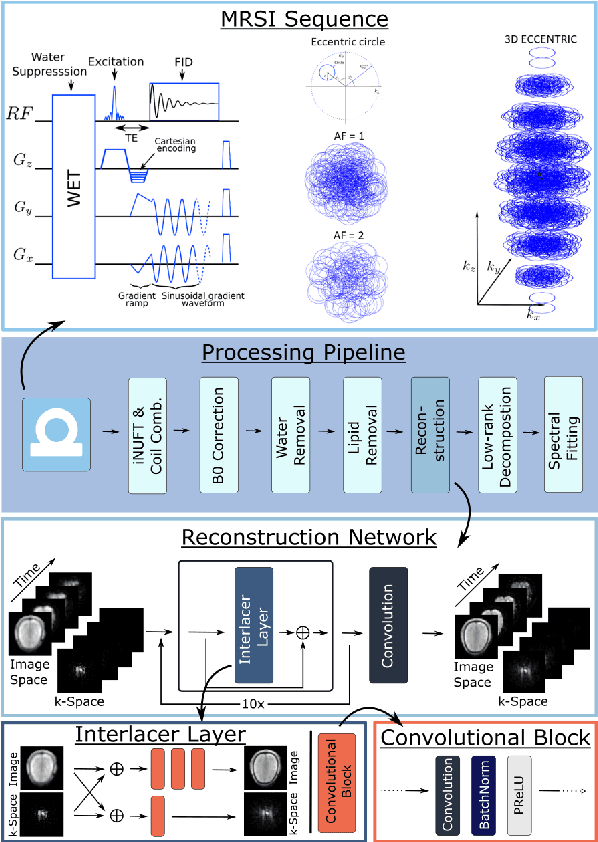
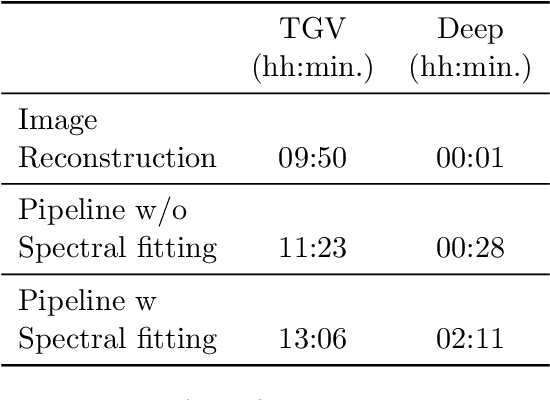

Abstract:Introduction: Altered neurometabolism is an important pathological mechanism in many neurological diseases and brain cancer, which can be mapped non-invasively by Magnetic Resonance Spectroscopic Imaging (MRSI). Advanced MRSI using non-cartesian compressed-sense acquisition enables fast high-resolution metabolic imaging but has lengthy reconstruction times that limits throughput and needs expert user interaction. Here, we present a robust and efficient Deep Learning reconstruction to obtain high-quality metabolic maps. Methods: Fast high-resolution whole-brain metabolic imaging was performed at 3.4 mm$^3$ isotropic resolution with acquisition times between 4:11-9:21 min:s using ECCENTRIC pulse sequence on a 7T MRI scanner. Data were acquired in a high-resolution phantom and 27 human participants, including 22 healthy volunteers and 5 glioma patients. A deep neural network using recurring interlaced convolutional layers with joint dual-space feature representation was developed for deep learning ECCENTRIC reconstruction (Deep-ER). 21 subjects were used for training and 6 subjects for testing. Deep-ER performance was compared to conventional iterative Total Generalized Variation reconstruction using image and spectral quality metrics. Results: Deep-ER demonstrated 600-fold faster reconstruction than conventional methods, providing improved spatial-spectral quality and metabolite quantification with 12%-45% (P<0.05) higher signal-to-noise and 8%-50% (P<0.05) smaller Cramer-Rao lower bounds. Metabolic images clearly visualize glioma tumor heterogeneity and boundary. Conclusion: Deep-ER provides efficient and robust reconstruction for sparse-sampled MRSI. The accelerated acquisition-reconstruction MRSI is compatible with high-throughput imaging workflow. It is expected that such improved performance will facilitate basic and clinical MRSI applications.
ECCENTRIC: a fast and unrestrained approach for high-resolution in vivo metabolic imaging at ultra-high field MR
May 23, 2023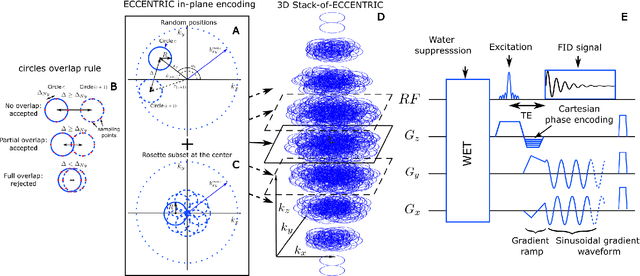
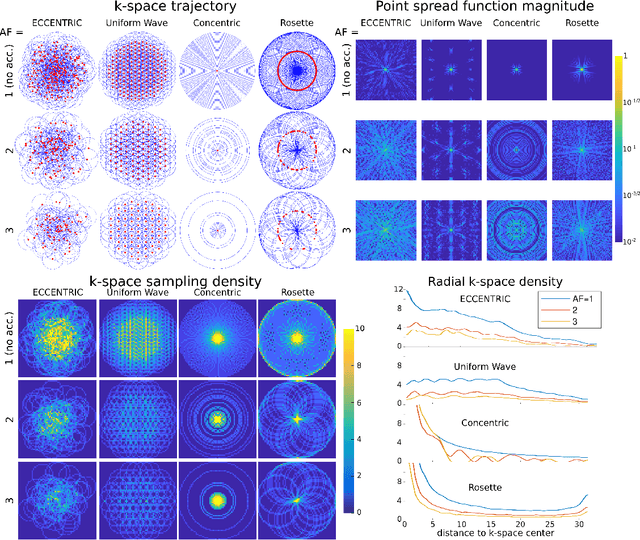
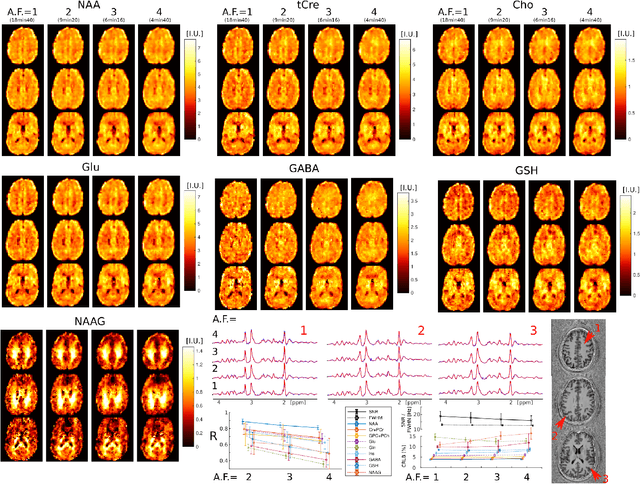
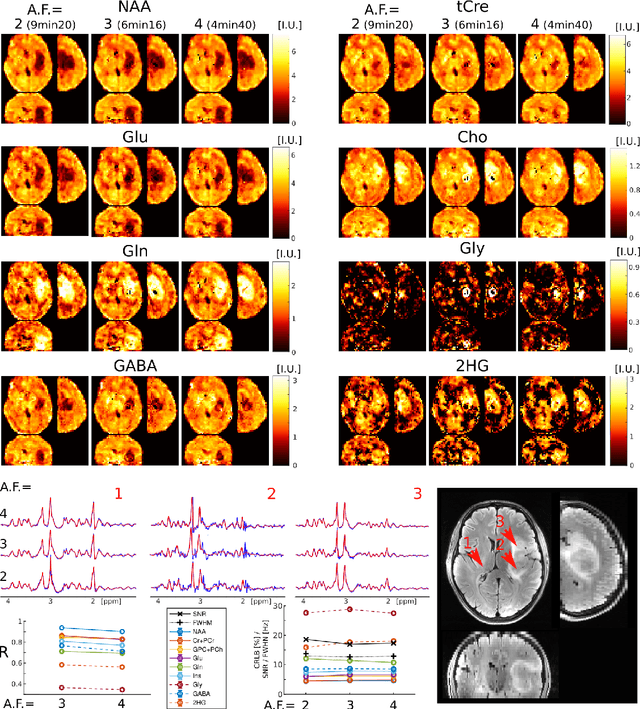
Abstract:A novel method for fast and high-resolution metabolic imaging, called ECcentric Circle ENcoding TRajectorIes for Compressed sensing (ECCENTRIC), has been developed and implemented on 7 Tesla human MRI. ECCENTRIC is a non-Cartesian spatial-spectral encoding method optimized for random undersampling of magnetic resonance spectroscopic imaging (MRSI) at ultra-high field. The approach provides flexible and random (k,t) sampling without temporal interleaving to improve spatial response function and spectral quality. ECCENTRIC needs low gradient amplitudes and slew-rates that reduces electrical, mechanical and thermal stress of the scanner hardware, and is robust to timing imperfection and eddy-current delays. Combined with a model-based low-rank reconstruction, this approach enables simultaneous imaging of up to 14 metabolites over the whole-brain at 2-3mm isotropic resolution in 4-10 minutes with high signal-to-noise ratio. In 20 healthy volunteers and 20 glioma patients ECCENTRIC demonstrated unprecedented mapping of fine structural details of metabolism in healthy brains and an extended metabolic fingerprinting of glioma tumors.
Predicting dynamic, motion-related changes in B0 field in the brain at a 7 T MRI using a subject-specific fine-tuned U-net
Apr 17, 2023Abstract:Subject movement during the magnetic resonance examination is inevitable and causes not only image artefacts but also deteriorates the homogeneity of the main magnetic field (B0), which is a prerequisite for high quality data. Thus, characterization of changes to B0, e.g. induced by patient movement, is important for MR applications that are prone to B0 inhomogeneities. We propose a deep learning based method to predict such changes within the brain from the change of the head position to facilitate retrospective or even real-time correction. A 3D U-net was trained on in vivo brain 7T MRI data. The input consisted of B0 maps and anatomical images at an initial position, and anatomical images at a different head position (obtained by applying a rigid-body transformation on the initial anatomical image). The output consisted of B0 maps at the new head positions. We further fine-tuned the network weights to each subject by measuring a limited number of head positions of the given subject, and trained the U-net with these data. Our approach was compared to established dynamic B0 field mapping via interleaved navigators, which suffer from limited spatial resolution and the need for undesirable sequence modifications. Qualitative and quantitative comparison showed similar performance between an interleaved navigator-equivalent method and proposed method. We therefore conclude that it is feasible to predict B0 maps from rigid subject movement and, when combined with external tracking hardware, this information could be used to improve the quality of magnetic resonance acquisitions without the use of navigators.
Flow-based Visual Quality Enhancer for Super-resolution Magnetic Resonance Spectroscopic Imaging
Jul 20, 2022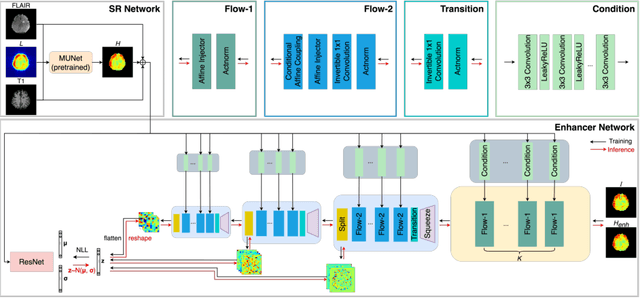
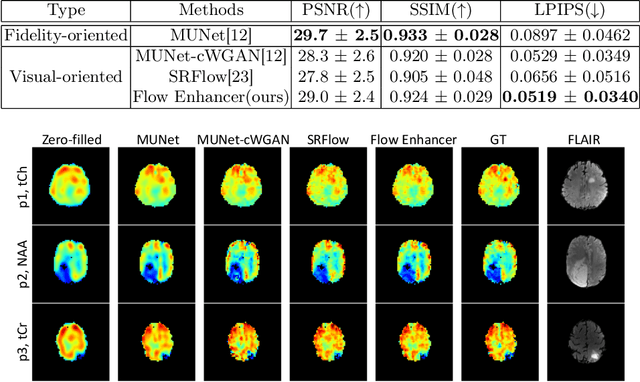
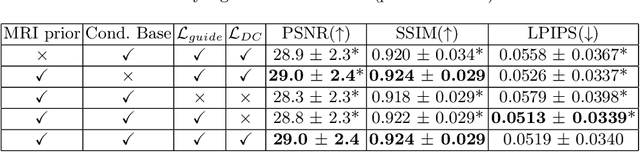
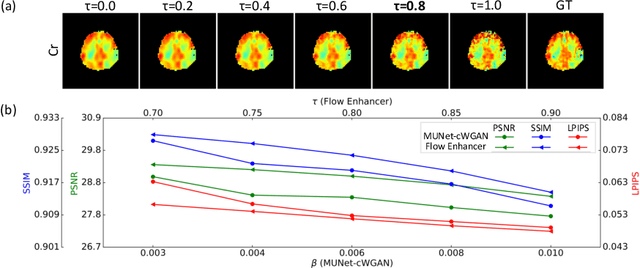
Abstract:Magnetic Resonance Spectroscopic Imaging (MRSI) is an essential tool for quantifying metabolites in the body, but the low spatial resolution limits its clinical applications. Deep learning-based super-resolution methods provided promising results for improving the spatial resolution of MRSI, but the super-resolved images are often blurry compared to the experimentally-acquired high-resolution images. Attempts have been made with the generative adversarial networks to improve the image visual quality. In this work, we consider another type of generative model, the flow-based model, of which the training is more stable and interpretable compared to the adversarial networks. Specifically, we propose a flow-based enhancer network to improve the visual quality of super-resolution MRSI. Different from previous flow-based models, our enhancer network incorporates anatomical information from additional image modalities (MRI) and uses a learnable base distribution. In addition, we impose a guide loss and a data-consistency loss to encourage the network to generate images with high visual quality while maintaining high fidelity. Experiments on a 1H-MRSI dataset acquired from 25 high-grade glioma patients indicate that our enhancer network outperforms the adversarial networks and the baseline flow-based methods. Our method also allows visual quality adjustment and uncertainty estimation.
Multi-scale Super-resolution Magnetic Resonance Spectroscopic Imaging with Adjustable Sharpness
Jun 17, 2022



Abstract:Magnetic Resonance Spectroscopic Imaging (MRSI) is a valuable tool for studying metabolic activities in the human body, but the current applications are limited to low spatial resolutions. The existing deep learning-based MRSI super-resolution methods require training a separate network for each upscaling factor, which is time-consuming and memory inefficient. We tackle this multi-scale super-resolution problem using a Filter Scaling strategy that modulates the convolution filters based on the upscaling factor, such that a single network can be used for various upscaling factors. Observing that each metabolite has distinct spatial characteristics, we also modulate the network based on the specific metabolite. Furthermore, our network is conditioned on the weight of adversarial loss so that the perceptual sharpness of the super-resolved metabolic maps can be adjusted within a single network. We incorporate these network conditionings using a novel Multi-Conditional Module. The experiments were carried out on a 1H-MRSI dataset from 15 high-grade glioma patients. Results indicate that the proposed network achieves the best performance among several multi-scale super-resolution methods and can provide super-resolved metabolic maps with adjustable sharpness.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge