WALINET: A water and lipid identification convolutional Neural Network for nuisance signal removal in 1H MR Spectroscopic Imaging
Paper and Code
Oct 01, 2024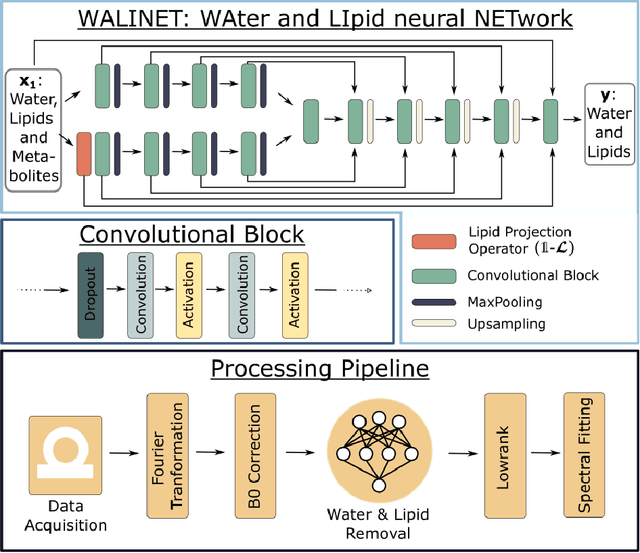

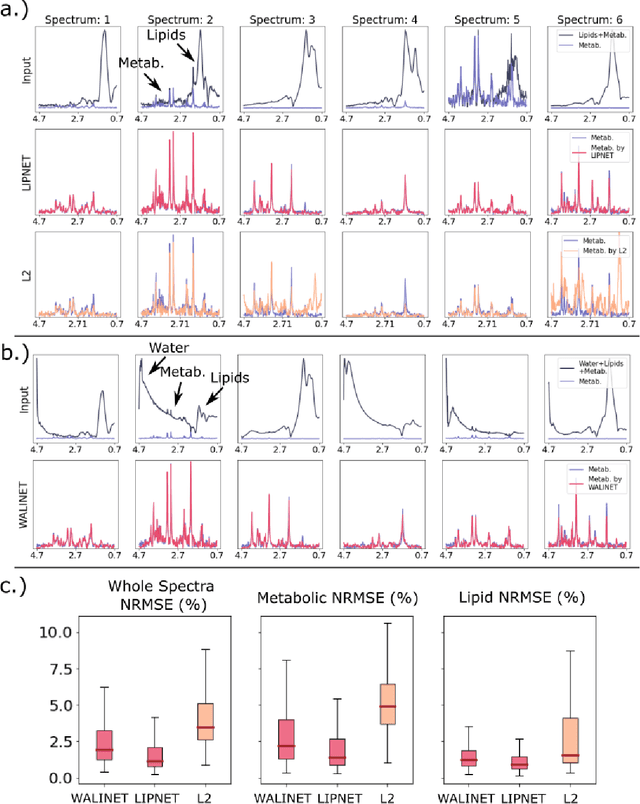
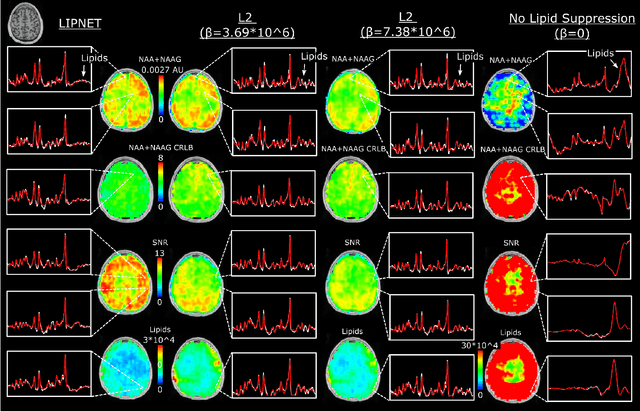
Purpose. Proton Magnetic Resonance Spectroscopic Imaging (1H-MRSI) provides non-invasive spectral-spatial mapping of metabolism. However, long-standing problems in whole-brain 1H-MRSI are spectral overlap of metabolite peaks with large lipid signal from scalp, and overwhelming water signal that distorts spectra. Fast and effective methods are needed for high-resolution 1H-MRSI to accurately remove lipid and water signals while preserving the metabolite signal. The potential of supervised neural networks for this task remains unexplored, despite their success for other MRSI processing. Methods. We introduce a deep-learning method based on a modified Y-NET network for water and lipid removal in whole-brain 1H-MRSI. The WALINET (WAter and LIpid neural NETwork) was compared to conventional methods such as the state-of-the-art lipid L2 regularization and Hankel-Lanczos singular value decomposition (HLSVD) water suppression. Methods were evaluated on simulated and in-vivo whole-brain MRSI using NMRSE, SNR, CRLB, and FWHM metrics. Results. WALINET is significantly faster and needs 8s for high-resolution whole-brain MRSI, compared to 42 minutes for conventional HLSVD+L2. Quantitative analysis shows WALINET has better performance than HLSVD+L2: 1) more lipid removal with 41% lower NRMSE, 2) better metabolite signal preservation with 71% lower NRMSE in simulated data, 155% higher SNR and 50% lower CRLB in in-vivo data. Metabolic maps obtained by WALINET in healthy subjects and patients show better gray/white-matter contrast with more visible structural details. Conclusions. WALINET has superior performance for nuisance signal removal and metabolite quantification on whole-brain 1H-MRSI compared to conventional state-of-the-art techniques. This represents a new application of deep-learning for MRSI processing, with potential for automated high-throughput workflow.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge