Daniel Ferreira
Synthesizing Individualized Aging Brains in Health and Disease with Generative Models and Parallel Transport
Feb 28, 2025



Abstract:Simulating prospective magnetic resonance imaging (MRI) scans from a given individual brain image is challenging, as it requires accounting for canonical changes in aging and/or disease progression while also considering the individual brain's current status and unique characteristics. While current deep generative models can produce high-resolution anatomically accurate templates for population-wide studies, their ability to predict future aging trajectories for individuals remains limited, particularly in capturing subject-specific neuroanatomical variations over time. In this study, we introduce Individualized Brain Synthesis (InBrainSyn), a framework for synthesizing high-resolution subject-specific longitudinal MRI scans that simulate neurodegeneration in both Alzheimer's disease (AD) and normal aging. InBrainSyn uses a parallel transport algorithm to adapt the population-level aging trajectories learned by a generative deep template network, enabling individualized aging synthesis. As InBrainSyn uses diffeomorphic transformations to simulate aging, the synthesized images are topologically consistent with the original anatomy by design. We evaluated InBrainSyn both quantitatively and qualitatively on AD and healthy control cohorts from the Open Access Series of Imaging Studies - version 3 dataset. Experimentally, InBrainSyn can also model neuroanatomical transitions between normal aging and AD. An evaluation of an external set supports its generalizability. Overall, with only a single baseline scan, InBrainSyn synthesizes realistic 3D spatiotemporal T1w MRI scans, producing personalized longitudinal aging trajectories. The code for InBrainSyn is available at: https://github.com/Fjr9516/InBrainSyn.
A deformation-based morphometry framework for disentangling Alzheimer's disease from normal aging using learned normal aging templates
Nov 14, 2023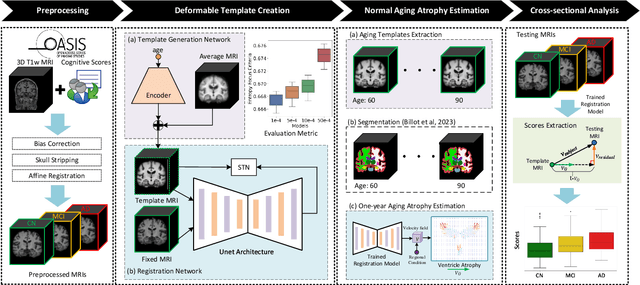
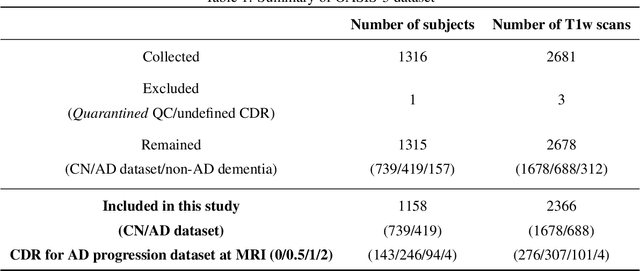
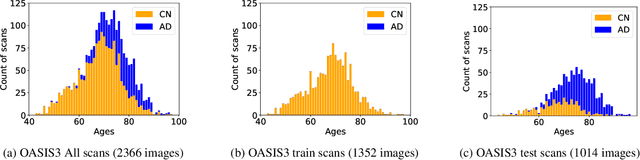
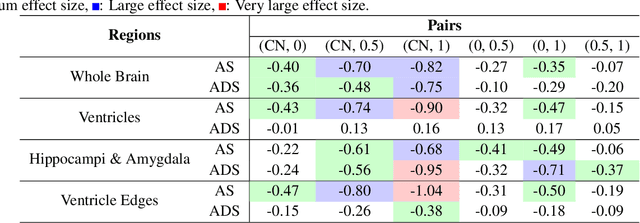
Abstract:Alzheimer's Disease and normal aging are both characterized by brain atrophy. The question of whether AD-related brain atrophy represents accelerated aging or a neurodegeneration process distinct from that in normal aging remains unresolved. Moreover, precisely disentangling AD-related brain atrophy from normal aging in a clinical context is complex. In this study, we propose a deformation-based morphometry framework to estimate normal aging and AD-specific atrophy patterns of subjects from morphological MRI scans. We first leverage deep-learning-based methods to create age-dependent templates of cognitively normal (CN) subjects. These templates model the normal aging atrophy patterns in a CN population. Then, we use the learned diffeomorphic registration to estimate the one-year normal aging pattern at the voxel level. We register the testing image to the 60-year-old CN template in the second step. Finally, normal aging and AD-specific scores are estimated by measuring the alignment of this registration with the one-year normal aging pattern. The methodology was developed and evaluated on the OASIS3 dataset with 1,014 T1-weighted MRI scans. Of these, 326 scans were from CN subjects, and 688 scans were from individuals clinically diagnosed with AD at different stages of clinical severity defined by clinical dementia rating (CDR) scores. The results show that ventricles predominantly follow an accelerated normal aging pattern in subjects with AD. In turn, hippocampi and amygdala regions were affected by both normal aging and AD-specific factors. Interestingly, hippocampi and amygdala regions showed more of an accelerated normal aging pattern for subjects during the early clinical stages of the disease, while the AD-specific score increases in later clinical stages. Our code is freely available at https://github.com/Fjr9516/DBM_with_DL.
Generative Aging of Brain Images with Diffeomorphic Registration
May 31, 2022



Abstract:Analyzing and predicting brain aging is essential for early prognosis and accurate diagnosis of cognitive diseases. The technique of neuroimaging, such as Magnetic Resonance Imaging (MRI), provides a noninvasive means of observing the aging process within the brain. With longitudinal image data collection, data-intensive Artificial Intelligence (AI) algorithms have been used to examine brain aging. However, existing state-of-the-art algorithms tend to be restricted to group-level predictions and suffer from unreal predictions. This paper proposes a methodology for generating longitudinal MRI scans that capture subject-specific neurodegeneration and retain anatomical plausibility in aging. The proposed methodology is developed within the framework of diffeomorphic registration and relies on three key novel technological advances to generate subject-level anatomically plausible predictions: i) a computationally efficient and individualized generative framework based on registration; ii) an aging generative module based on biological linear aging progression; iii) a quality control module to fit registration for generation task. Our methodology was evaluated on 2662 T1-weighted (T1-w) MRI scans from 796 participants from three different cohorts. First, we applied 6 commonly used criteria to demonstrate the aging simulation ability of the proposed methodology; Secondly, we evaluated the quality of the synthetic images using quantitative measurements and qualitative assessment by a neuroradiologist. Overall, the experimental results show that the proposed method can produce anatomically plausible predictions that can be used to enhance longitudinal datasets, in turn enabling data-hungry AI-driven healthcare tools.
The reliability of a deep learning model in clinical out-of-distribution MRI data: a multicohort study
Nov 01, 2019



Abstract:Deep learning (DL) methods have in recent years yielded impressive results in medical imaging, with the potential to function as clinical aid to radiologists. However, DL models in medical imaging are often trained on public research cohorts with images acquired with a single scanner or with strict protocol harmonization, which is not representative of a clinical setting. The aim of this study was to investigate how well a DL model performs in unseen clinical data sets---collected with different scanners, protocols and disease populations---and whether more heterogeneous training data improves generalization. In total, 3117 MRI scans of brains from multiple dementia research cohorts and memory clinics, that had been visually rated by a neuroradiologist according to Scheltens' scale of medial temporal atrophy (MTA), were included in this study. By training multiple versions of a convolutional neural network on different subsets of this data to predict MTA ratings, we assessed the impact of including images from a wider distribution during training had on performance in external memory clinic data. Our results showed that our model generalized well to data sets acquired with similar protocols as the training data, but substantially worse in clinical cohorts with visibly different tissue contrasts in the images. This implies that future DL studies investigating performance in out-of-distribution (OOD) MRI data need to assess multiple external cohorts for reliable results. Further, by including data from a wider range of scanners and protocols the performance improved in OOD data, which suggests that more heterogeneous training data makes the model generalize better. To conclude, this is the most comprehensive study to date investigating the domain shift in deep learning on MRI data, and we advocate rigorous evaluation of DL models on clinical data prior to being certified for deployment.
Towards modular and programmable architecture search
Sep 30, 2019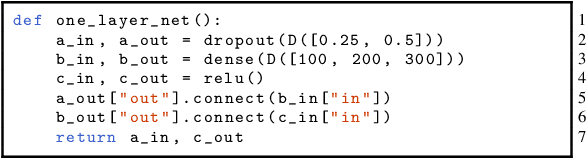
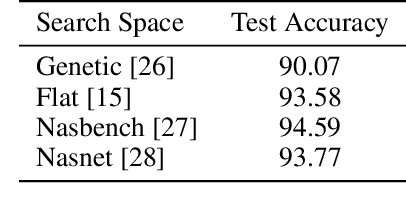

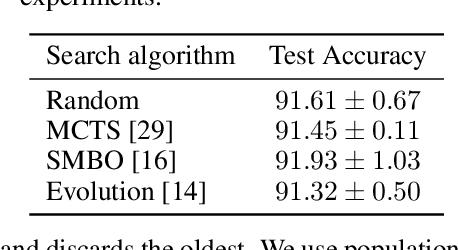
Abstract:Neural architecture search methods are able to find high performance deep learning architectures with minimal effort from an expert. However, current systems focus on specific use-cases (e.g. convolutional image classifiers and recurrent language models), making them unsuitable for general use-cases that an expert might wish to write. Hyperparameter optimization systems are general-purpose but lack the constructs needed for easy application to architecture search. In this work, we propose a formal language for encoding search spaces over general computational graphs. The language constructs allow us to write modular, composable, and reusable search space encodings and to reason about search space design. We use our language to encode search spaces from the architecture search literature. The language allows us to decouple the implementations of the search space and the search algorithm, allowing us to expose search spaces to search algorithms through a consistent interface. Our experiments show the ease with which we can experiment with different combinations of search spaces and search algorithms without having to implement each combination from scratch. We release an implementation of our language with this paper.
AVRA: Automatic Visual Ratings of Atrophy from MRI images using Recurrent Convolutional Neural Networks
Dec 23, 2018



Abstract:Quantifying the degree of atrophy is done clinically by neuroradiologists following established visual rating scales. For these assessments to be reliable the rater requires substantial training and experience, and even then the rating agreement between two radiologists is not perfect. We have developed a model we call AVRA (Automatic Visual Ratings of Atrophy) based on machine learning methods and trained on 2350 visual ratings made by an experienced neuroradiologist. It provides fast and automatic ratings for Scheltens' scale of medial temporal atrophy (MTA), the frontal subscale of Pasquier's Global Cortical Atrophy (GCA-F) scale, and Koedam's scale of Posterior Atrophy (PA). We demonstrate substantial inter-rater agreement between AVRA's and a neuroradiologist ratings with Cohen's weighted kappa values of $\kappa_w$ = 0.74/0.72 (MTA left/right), $\kappa_w$ = 0.62 (GCA-F) and $\kappa_w$ = 0.74 (PA), with an inherent intra-rater agreement of $\kappa_w$ = 1. We conclude that automatic visual ratings of atrophy can potentially have great clinical and scientific value, and aim to present AVRA as a freely available toolbox.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge