A deformation-based morphometry framework for disentangling Alzheimer's disease from normal aging using learned normal aging templates
Paper and Code
Nov 14, 2023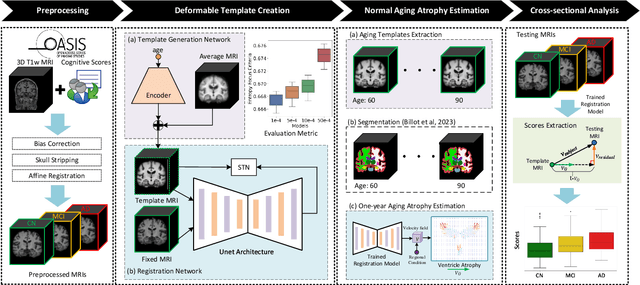
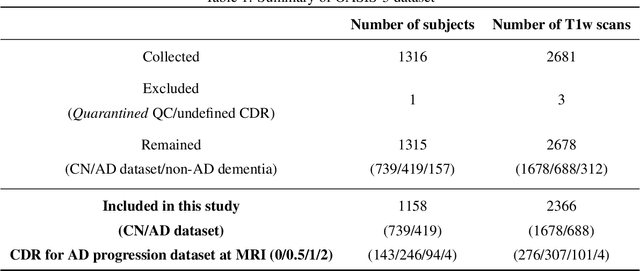
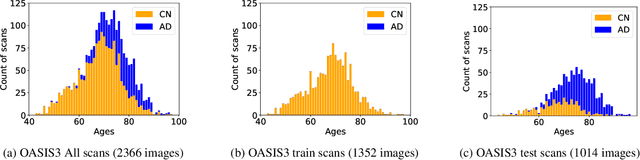
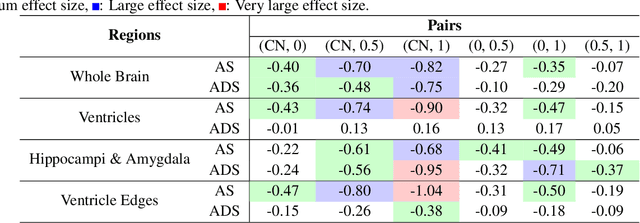
Alzheimer's Disease and normal aging are both characterized by brain atrophy. The question of whether AD-related brain atrophy represents accelerated aging or a neurodegeneration process distinct from that in normal aging remains unresolved. Moreover, precisely disentangling AD-related brain atrophy from normal aging in a clinical context is complex. In this study, we propose a deformation-based morphometry framework to estimate normal aging and AD-specific atrophy patterns of subjects from morphological MRI scans. We first leverage deep-learning-based methods to create age-dependent templates of cognitively normal (CN) subjects. These templates model the normal aging atrophy patterns in a CN population. Then, we use the learned diffeomorphic registration to estimate the one-year normal aging pattern at the voxel level. We register the testing image to the 60-year-old CN template in the second step. Finally, normal aging and AD-specific scores are estimated by measuring the alignment of this registration with the one-year normal aging pattern. The methodology was developed and evaluated on the OASIS3 dataset with 1,014 T1-weighted MRI scans. Of these, 326 scans were from CN subjects, and 688 scans were from individuals clinically diagnosed with AD at different stages of clinical severity defined by clinical dementia rating (CDR) scores. The results show that ventricles predominantly follow an accelerated normal aging pattern in subjects with AD. In turn, hippocampi and amygdala regions were affected by both normal aging and AD-specific factors. Interestingly, hippocampi and amygdala regions showed more of an accelerated normal aging pattern for subjects during the early clinical stages of the disease, while the AD-specific score increases in later clinical stages. Our code is freely available at https://github.com/Fjr9516/DBM_with_DL.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge