Ketil Oppedal
Out-of-distribution multi-view auto-encoders for prostate cancer lesion detection
Aug 12, 2023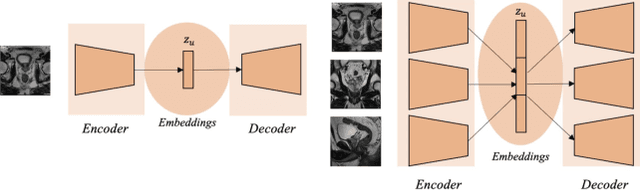
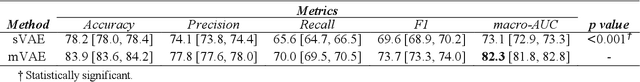
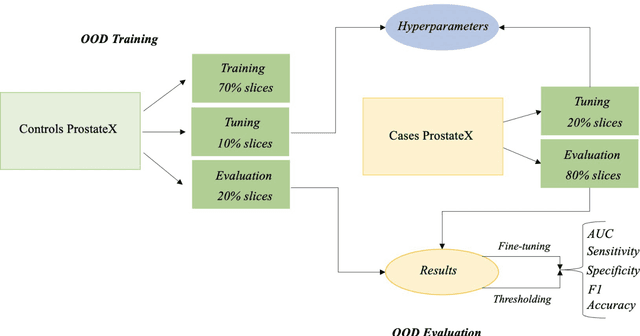
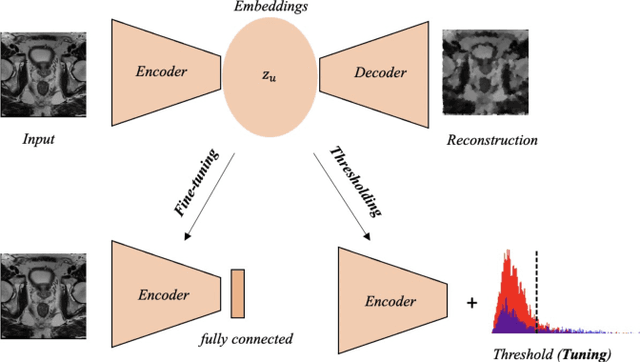
Abstract:Traditional deep learning (DL) approaches based on supervised learning paradigms require large amounts of annotated data that are rarely available in the medical domain. Unsupervised Out-of-distribution (OOD) detection is an alternative that requires less annotated data. Further, OOD applications exploit the class skewness commonly present in medical data. Magnetic resonance imaging (MRI) has proven to be useful for prostate cancer (PCa) diagnosis and management, but current DL approaches rely on T2w axial MRI, which suffers from low out-of-plane resolution. We propose a multi-stream approach to accommodate different T2w directions to improve the performance of PCa lesion detection in an OOD approach. We evaluate our approach on a publicly available data-set, obtaining better detection results in terms of AUC when compared to a single direction approach (73.1 vs 82.3). Our results show the potential of OOD approaches for PCa lesion detection based on MRI.
3D Masked Modelling Advances Lesion Classification in Axial T2w Prostate MRI
Dec 29, 2022Abstract:Masked Image Modelling (MIM) has been shown to be an efficient self-supervised learning (SSL) pre-training paradigm when paired with transformer architectures and in the presence of a large amount of unlabelled natural images. The combination of the difficulties in accessing and obtaining large amounts of labeled data and the availability of unlabelled data in the medical imaging domain makes MIM an interesting approach to advance deep learning (DL) applications based on 3D medical imaging data. Nevertheless, SSL and, in particular, MIM applications with medical imaging data are rather scarce and there is still uncertainty. around the potential of such a learning paradigm in the medical domain. We study MIM in the context of Prostate Cancer (PCa) lesion classification with T2 weighted (T2w) axial magnetic resonance imaging (MRI) data. In particular, we explore the effect of using MIM when coupled with convolutional neural networks (CNNs) under different conditions such as different masking strategies, obtaining better results in terms of AUC than other pre-training strategies like ImageNet weight initialization.
Self-transfer learning via patches: A prostate cancer triage approach based on bi-parametric MRI
Jul 22, 2021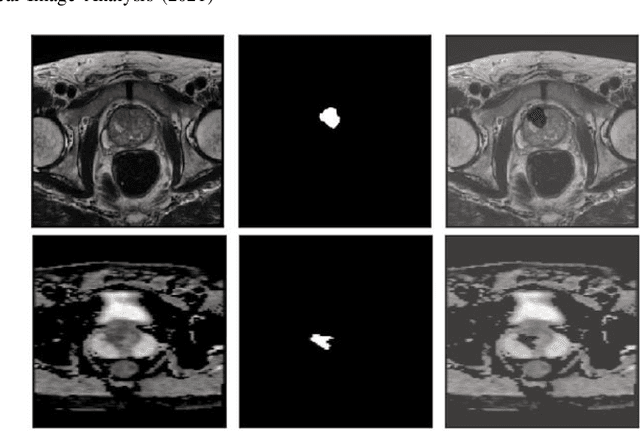
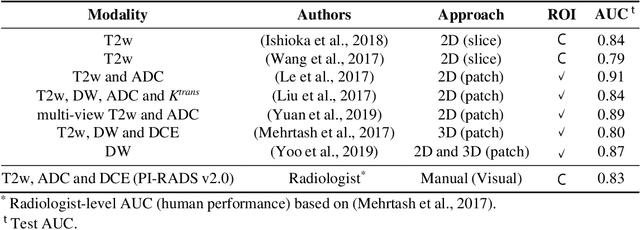
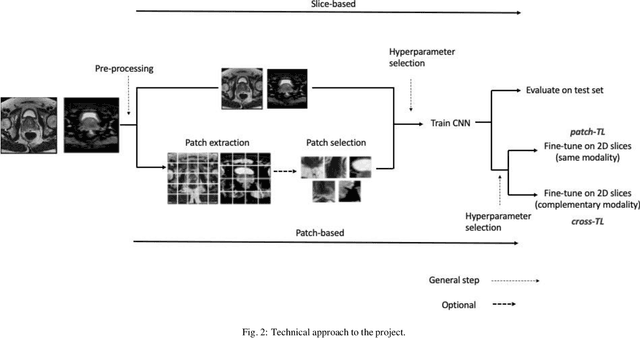
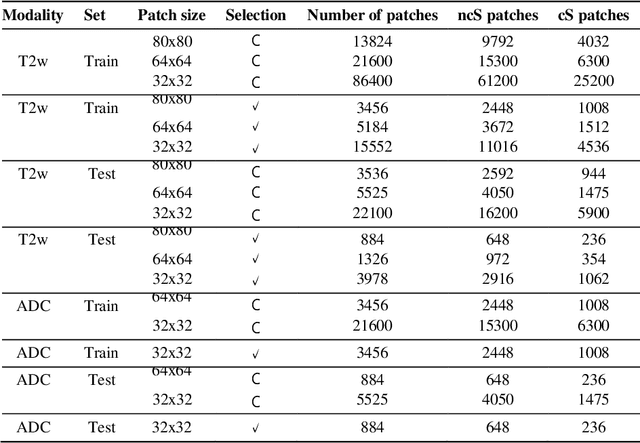
Abstract:Prostate cancer (PCa) is the second most common cancer diagnosed among men worldwide. The current PCa diagnostic pathway comes at the cost of substantial overdiagnosis, leading to unnecessary treatment and further testing. Bi-parametric magnetic resonance imaging (bp-MRI) based on apparent diffusion coefficient maps (ADC) and T2-weighted (T2w) sequences has been proposed as a triage test to differentiate between clinically significant (cS) and non-clinically significant (ncS) prostate lesions. However, analysis of the sequences relies on expertise, requires specialized training, and suffers from inter-observer variability. Deep learning (DL) techniques hold promise in tasks such as classification and detection. Nevertheless, they rely on large amounts of annotated data which is not common in the medical field. In order to palliate such issues, existing works rely on transfer learning (TL) and ImageNet pre-training, which has been proven to be sub-optimal for the medical imaging domain. In this paper, we present a patch-based pre-training strategy to distinguish between cS and ncS lesions which exploit the region of interest (ROI) of the patched source domain to efficiently train a classifier in the full-slice target domain which does not require annotations by making use of transfer learning (TL). We provide a comprehensive comparison between several CNNs architectures and different settings which are presented as a baseline. Moreover, we explore cross-domain TL which exploits both MRI modalities and improves single modality results. Finally, we show how our approaches outperform the standard approaches by a considerable margin
Improving prostate whole gland segmentation in t2-weighted MRI with synthetically generated data
Mar 27, 2021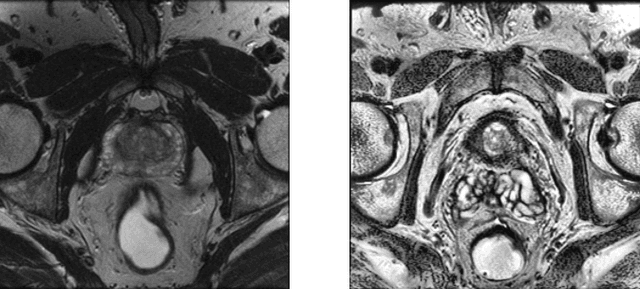

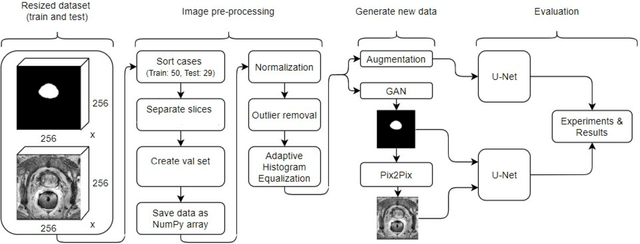

Abstract:Whole gland (WG) segmentation of the prostate plays a crucial role in detection, staging and treatment planning of prostate cancer (PCa). Despite promise shown by deep learning (DL) methods, they rely on the availability of a considerable amount of annotated data. Augmentation techniques such as translation and rotation of images present an alternative to increase data availability. Nevertheless, the amount of information provided by the transformed data is limited due to the correlation between the generated data and the original. Based on the recent success of generative adversarial networks (GAN) in producing synthetic images for other domains as well as in the medical domain, we present a pipeline to generate WG segmentation masks and synthesize T2-weighted MRI of the prostate based on a publicly available multi-center dataset. Following, we use the generated data as a form of data augmentation. Results show an improvement in the quality of the WG segmentation when compared to standard augmentation techniques.
The reliability of a deep learning model in clinical out-of-distribution MRI data: a multicohort study
Nov 01, 2019



Abstract:Deep learning (DL) methods have in recent years yielded impressive results in medical imaging, with the potential to function as clinical aid to radiologists. However, DL models in medical imaging are often trained on public research cohorts with images acquired with a single scanner or with strict protocol harmonization, which is not representative of a clinical setting. The aim of this study was to investigate how well a DL model performs in unseen clinical data sets---collected with different scanners, protocols and disease populations---and whether more heterogeneous training data improves generalization. In total, 3117 MRI scans of brains from multiple dementia research cohorts and memory clinics, that had been visually rated by a neuroradiologist according to Scheltens' scale of medial temporal atrophy (MTA), were included in this study. By training multiple versions of a convolutional neural network on different subsets of this data to predict MTA ratings, we assessed the impact of including images from a wider distribution during training had on performance in external memory clinic data. Our results showed that our model generalized well to data sets acquired with similar protocols as the training data, but substantially worse in clinical cohorts with visibly different tissue contrasts in the images. This implies that future DL studies investigating performance in out-of-distribution (OOD) MRI data need to assess multiple external cohorts for reliable results. Further, by including data from a wider range of scanners and protocols the performance improved in OOD data, which suggests that more heterogeneous training data makes the model generalize better. To conclude, this is the most comprehensive study to date investigating the domain shift in deep learning on MRI data, and we advocate rigorous evaluation of DL models on clinical data prior to being certified for deployment.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge