Trygve Eftestøl
NMGrad: Advancing Histopathological Bladder Cancer Grading with Weakly Supervised Deep Learning
May 24, 2024Abstract:The most prevalent form of bladder cancer is urothelial carcinoma, characterized by a high recurrence rate and substantial lifetime treatment costs for patients. Grading is a prime factor for patient risk stratification, although it suffers from inconsistencies and variations among pathologists. Moreover, absence of annotations in medical imaging difficults training deep learning models. To address these challenges, we introduce a pipeline designed for bladder cancer grading using histological slides. First, it extracts urothelium tissue tiles at different magnification levels, employing a convolutional neural network for processing for feature extraction. Then, it engages in the slide-level prediction process. It employs a nested multiple instance learning approach with attention to predict the grade. To distinguish different levels of malignancy within specific regions of the slide, we include the origins of the tiles in our analysis. The attention scores at region level is shown to correlate with verified high-grade regions, giving some explainability to the model. Clinical evaluations demonstrate that our model consistently outperforms previous state-of-the-art methods.
Self-Contrastive Weakly Supervised Learning Framework for Prognostic Prediction Using Whole Slide Images
May 24, 2024



Abstract:We present a pioneering investigation into the application of deep learning techniques to analyze histopathological images for addressing the substantial challenge of automated prognostic prediction. Prognostic prediction poses a unique challenge as the ground truth labels are inherently weak, and the model must anticipate future events that are not directly observable in the image. To address this challenge, we propose a novel three-part framework comprising of a convolutional network based tissue segmentation algorithm for region of interest delineation, a contrastive learning module for feature extraction, and a nested multiple instance learning classification module. Our study explores the significance of various regions of interest within the histopathological slides and exploits diverse learning scenarios. The pipeline is initially validated on artificially generated data and a simpler diagnostic task. Transitioning to prognostic prediction, tasks become more challenging. Employing bladder cancer as use case, our best models yield an AUC of 0.721 and 0.678 for recurrence and treatment outcome prediction respectively.
Reconsidering evaluation practices in modular systems: On the propagation of errors in MRI prostate cancer detection
Sep 15, 2023Abstract:Magnetic resonance imaging has evolved as a key component for prostate cancer (PCa) detection, substantially increasing the radiologist workload. Artificial intelligence (AI) systems can support radiological assessment by segmenting and classifying lesions in clinically significant (csPCa) and non-clinically significant (ncsPCa). Commonly, AI systems for PCa detection involve an automatic prostate segmentation followed by the lesion detection using the extracted prostate. However, evaluation reports are typically presented in terms of detection under the assumption of the availability of a highly accurate segmentation and an idealistic scenario, omitting the propagation of errors between modules. For that purpose, we evaluate the effect of two different segmentation networks (s1 and s2) with heterogeneous performances in the detection stage and compare it with an idealistic setting (s1:89.90+-2.23 vs 88.97+-3.06 ncsPCa, P<.001, 89.30+-4.07 and 88.12+-2.71 csPCa, P<.001). Our results depict the relevance of a holistic evaluation, accounting for all the sub-modules involved in the system.
Out-of-distribution multi-view auto-encoders for prostate cancer lesion detection
Aug 12, 2023
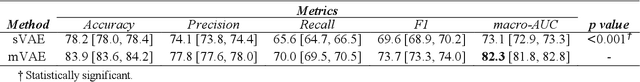
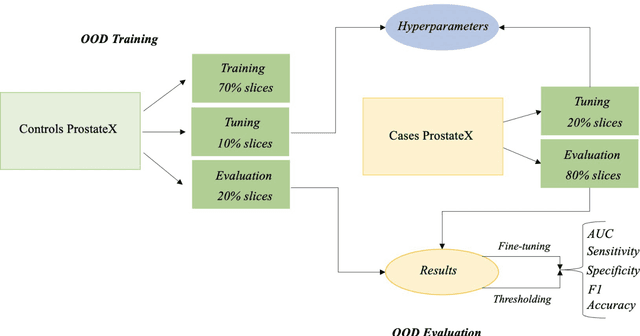
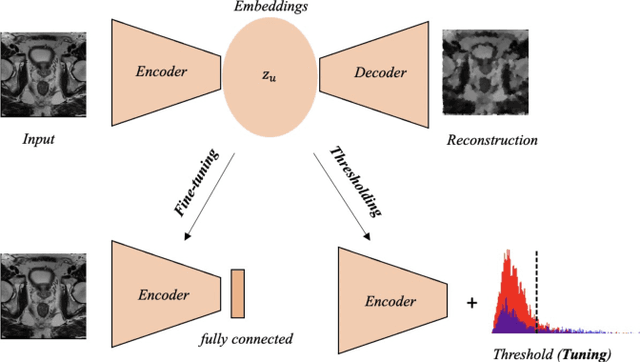
Abstract:Traditional deep learning (DL) approaches based on supervised learning paradigms require large amounts of annotated data that are rarely available in the medical domain. Unsupervised Out-of-distribution (OOD) detection is an alternative that requires less annotated data. Further, OOD applications exploit the class skewness commonly present in medical data. Magnetic resonance imaging (MRI) has proven to be useful for prostate cancer (PCa) diagnosis and management, but current DL approaches rely on T2w axial MRI, which suffers from low out-of-plane resolution. We propose a multi-stream approach to accommodate different T2w directions to improve the performance of PCa lesion detection in an OOD approach. We evaluate our approach on a publicly available data-set, obtaining better detection results in terms of AUC when compared to a single direction approach (73.1 vs 82.3). Our results show the potential of OOD approaches for PCa lesion detection based on MRI.
Leveraging multi-view data without annotations for prostate MRI segmentation: A contrastive approach
Aug 12, 2023Abstract:An accurate prostate delineation and volume characterization can support the clinical assessment of prostate cancer. A large amount of automatic prostate segmentation tools consider exclusively the axial MRI direction in spite of the availability as per acquisition protocols of multi-view data. Further, when multi-view data is exploited, manual annotations and availability at test time for all the views is commonly assumed. In this work, we explore a contrastive approach at training time to leverage multi-view data without annotations and provide flexibility at deployment time in the event of missing views. We propose a triplet encoder and single decoder network based on U-Net, tU-Net (triplet U-Net). Our proposed architecture is able to exploit non-annotated sagittal and coronal views via contrastive learning to improve the segmentation from a volumetric perspective. For that purpose, we introduce the concept of inter-view similarity in the latent space. To guide the training, we combine a dice score loss calculated with respect to the axial view and its manual annotations together with a multi-view contrastive loss. tU-Net shows statistical improvement in dice score coefficient (DSC) with respect to only axial view (91.25+-0.52% compared to 86.40+-1.50%,P<.001). Sensitivity analysis reveals the volumetric positive impact of the contrastive loss when paired with tU-Net (2.85+-1.34% compared to 3.81+-1.88%,P<.001). Further, our approach shows good external volumetric generalization in an in-house dataset when tested with multi-view data (2.76+-1.89% compared to 3.92+-3.31%,P=.002), showing the feasibility of exploiting non-annotated multi-view data through contrastive learning whilst providing flexibility at deployment in the event of missing views.
Activity Recognition From Newborn Resuscitation Videos
Mar 14, 2023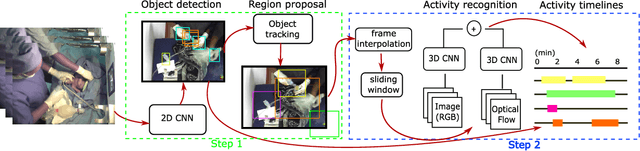
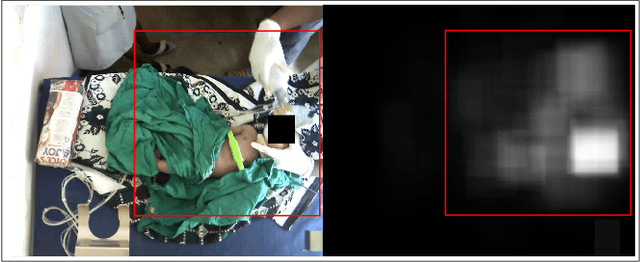
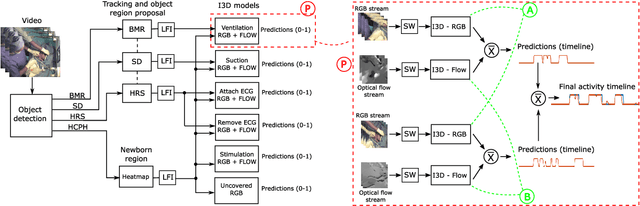
Abstract:Objective: Birth asphyxia is one of the leading causes of neonatal deaths. A key for survival is performing immediate and continuous quality newborn resuscitation. A dataset of recorded signals during newborn resuscitation, including videos, has been collected in Haydom, Tanzania, and the aim is to analyze the treatment and its effect on the newborn outcome. An important step is to generate timelines of relevant resuscitation activities, including ventilation, stimulation, suction, etc., during the resuscitation episodes. Methods: We propose a two-step deep neural network system, ORAA-net, utilizing low-quality video recordings of resuscitation episodes to do activity recognition during newborn resuscitation. The first step is to detect and track relevant objects using Convolutional Neural Networks (CNN) and post-processing, and the second step is to analyze the proposed activity regions from step 1 to do activity recognition using 3D CNNs. Results: The system recognized the activities newborn uncovered, stimulation, ventilation and suction with a mean precision of 77.67 %, a mean recall of 77,64 %, and a mean accuracy of 92.40 %. Moreover, the accuracy of the estimated number of Health Care Providers (HCPs) present during the resuscitation episodes was 68.32 %. Conclusion: The results indicate that the proposed CNN-based two-step ORAAnet could be used for object detection and activity recognition in noisy low-quality newborn resuscitation videos. Significance: A thorough analysis of the effect the different resuscitation activities have on the newborn outcome could potentially allow us to optimize treatment guidelines, training, debriefing, and local quality improvement in newborn resuscitation.
* 10 pages
Object Detection During Newborn Resuscitation Activities
Mar 14, 2023Abstract:Birth asphyxia is a major newborn mortality problem in low-resource countries. International guideline provides treatment recommendations; however, the importance and effect of the different treatments are not fully explored. The available data is collected in Tanzania, during newborn resuscitation, for analysis of the resuscitation activities and the response of the newborn. An important step in the analysis is to create activity timelines of the episodes, where activities include ventilation, suction, stimulation etc. Methods: The available recordings are noisy real-world videos with large variations. We propose a two-step process in order to detect activities possibly overlapping in time. The first step is to detect and track the relevant objects, like bag-mask resuscitator, heart rate sensors etc., and the second step is to use this information to recognize the resuscitation activities. The topic of this paper is the first step, and the object detection and tracking are based on convolutional neural networks followed by post processing. Results: The performance of the object detection during activities were 96.97 % (ventilations), 100 % (attaching/removing heart rate sensor) and 75 % (suction) on a test set of 20 videos. The system also estimate the number of health care providers present with a performance of 71.16 %. Conclusion: The proposed object detection and tracking system provides promising results in noisy newborn resuscitation videos. Significance: This is the first step in a thorough analysis of newborn resuscitation episodes, which could provide important insight about the importance and effect of different newborn resuscitation activities
* 8 pages
Active Learning Based Domain Adaptation for Tissue Segmentation of Histopathological Images
Mar 09, 2023Abstract:Accurate segmentation of tissue in histopathological images can be very beneficial for defining regions of interest (ROI) for streamline of diagnostic and prognostic tasks. Still, adapting to different domains is essential for histopathology image analysis, as the visual characteristics of tissues can vary significantly across datasets. Yet, acquiring sufficient annotated data in the medical domain is cumbersome and time-consuming. The labeling effort can be significantly reduced by leveraging active learning, which enables the selective annotation of the most informative samples. Our proposed method allows for fine-tuning a pre-trained deep neural network using a small set of labeled data from the target domain, while also actively selecting the most informative samples to label next. We demonstrate that our approach performs with significantly fewer labeled samples compared to traditional supervised learning approaches for similar F1-scores, using barely a 59\% of the training set. We also investigate the distribution of class balance to establish annotation guidelines.
3D Masked Modelling Advances Lesion Classification in Axial T2w Prostate MRI
Dec 29, 2022Abstract:Masked Image Modelling (MIM) has been shown to be an efficient self-supervised learning (SSL) pre-training paradigm when paired with transformer architectures and in the presence of a large amount of unlabelled natural images. The combination of the difficulties in accessing and obtaining large amounts of labeled data and the availability of unlabelled data in the medical imaging domain makes MIM an interesting approach to advance deep learning (DL) applications based on 3D medical imaging data. Nevertheless, SSL and, in particular, MIM applications with medical imaging data are rather scarce and there is still uncertainty. around the potential of such a learning paradigm in the medical domain. We study MIM in the context of Prostate Cancer (PCa) lesion classification with T2 weighted (T2w) axial magnetic resonance imaging (MRI) data. In particular, we explore the effect of using MIM when coupled with convolutional neural networks (CNNs) under different conditions such as different masking strategies, obtaining better results in terms of AUC than other pre-training strategies like ImageNet weight initialization.
Nested Multiple Instance Learning with Attention Mechanisms
Nov 02, 2021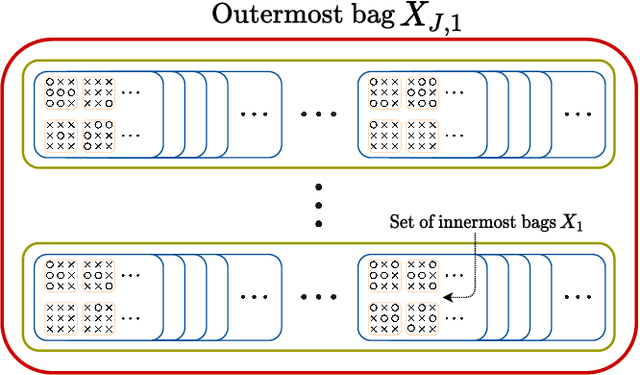
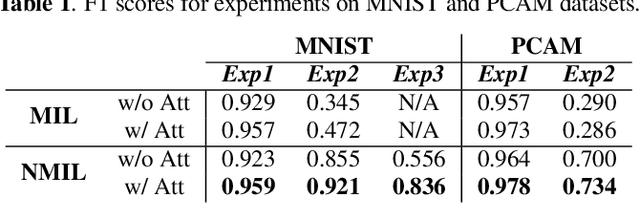
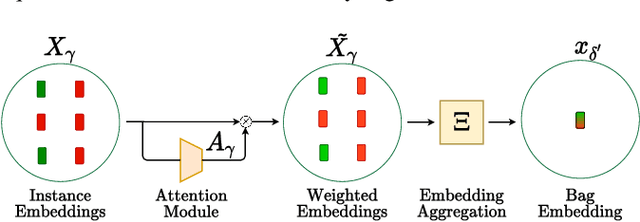
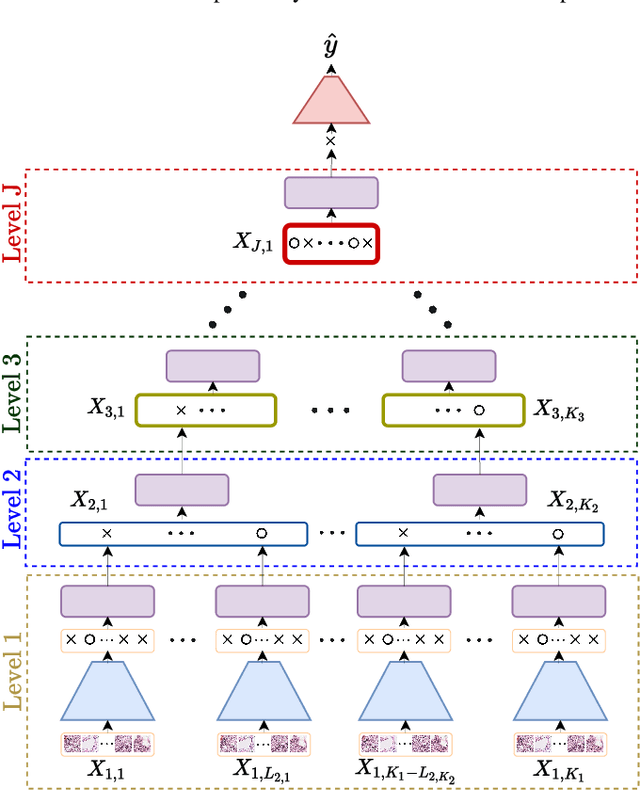
Abstract:Multiple instance learning (MIL) is a type of weakly supervised learning where multiple instances of data with unknown labels are sorted into bags. Since knowledge about the individual instances is incomplete, labels are assigned to the bags containing the instances. While this method fits diverse applications were labelled data is scarce, it lacks depth for solving more complex scenarios where associations between sets of instances have to be made, like finding relevant regions of interest in an image or detecting events in a set of time-series signals. Nested MIL considers labelled bags within bags, where only the outermost bag is labelled and inner-bags and instances are represented as latent labels. In addition, we propose using an attention mechanism to add interpretability, providing awareness into the impact of each instance to the weak bag label. Experiments in classical image datasets show that our proposed model provides high accuracy performance as well as spotting relevant instances on image regions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge