Valery Naranjo
HUMAN-tech Institute, Universitat Politènica de València, Valencia, Spain
Defect Segmentation in OCT scans of ceramic parts for non-destructive inspection using deep learning
Oct 01, 2025



Abstract:Non-destructive testing (NDT) is essential in ceramic manufacturing to ensure the quality of components without compromising their integrity. In this context, Optical Coherence Tomography (OCT) enables high-resolution internal imaging, revealing defects such as pores, delaminations, or inclusions. This paper presents an automatic defect detection system based on Deep Learning (DL), trained on OCT images with manually segmented annotations. A neural network based on the U-Net architecture is developed, evaluating multiple experimental configurations to enhance its performance. Post-processing techniques enable both quantitative and qualitative evaluation of the predictions. The system shows an accurate behavior of 0.979 Dice Score, outperforming comparable studies. The inference time of 18.98 seconds per volume supports its viability for detecting inclusions, enabling more efficient, reliable, and automated quality control.
Exploring visual language models as a powerful tool in the diagnosis of Ewing Sarcoma
Jan 14, 2025



Abstract:Ewing's sarcoma (ES), characterized by a high density of small round blue cells without structural organization, presents a significant health concern, particularly among adolescents aged 10 to 19. Artificial intelligence-based systems for automated analysis of histopathological images are promising to contribute to an accurate diagnosis of ES. In this context, this study explores the feature extraction ability of different pre-training strategies for distinguishing ES from other soft tissue or bone sarcomas with similar morphology in digitized tissue microarrays for the first time, as far as we know. Vision-language supervision (VLS) is compared to fully-supervised ImageNet pre-training within a multiple instance learning paradigm. Our findings indicate a substantial improvement in diagnostic accuracy with the adaption of VLS using an in-domain dataset. Notably, these models not only enhance the accuracy of predicted classes but also drastically reduce the number of trainable parameters and computational costs.
Enhancing Whole Slide Image Classification through Supervised Contrastive Domain Adaptation
Dec 05, 2024



Abstract:Domain shift in the field of histopathological imaging is a common phenomenon due to the intra- and inter-hospital variability of staining and digitization protocols. The implementation of robust models, capable of creating generalized domains, represents a need to be solved. In this work, a new domain adaptation method to deal with the variability between histopathological images from multiple centers is presented. In particular, our method adds a training constraint to the supervised contrastive learning approach to achieve domain adaptation and improve inter-class separability. Experiments performed on domain adaptation and classification of whole-slide images of six skin cancer subtypes from two centers demonstrate the method's usefulness. The results reflect superior performance compared to not using domain adaptation after feature extraction or staining normalization.
MI-VisionShot: Few-shot adaptation of vision-language models for slide-level classification of histopathological images
Oct 21, 2024



Abstract:Vision-language supervision has made remarkable strides in learning visual representations from textual guidance. In digital pathology, vision-language models (VLM), pre-trained on curated datasets of histological image-captions, have been adapted to downstream tasks, such as region of interest classification. Zero-shot transfer for slide-level prediction has been formulated by MI-Zero, but it exhibits high variability depending on the textual prompts. Inspired by prototypical learning, we propose MI-VisionShot, a training-free adaptation method on top of VLMs to predict slide-level labels in few-shot learning scenarios. Our framework takes advantage of the excellent representation learning of VLM to create prototype-based classifiers under a multiple-instance setting by retrieving the most discriminative patches within each slide. Experimentation through different settings shows the ability of MI-VisionShot to surpass zero-shot transfer with lower variability, even in low-shot scenarios. Code coming soon at thttps://github.com/cvblab/MIVisionShot.
Foundation Models for Slide-level Cancer Subtyping in Digital Pathology
Oct 21, 2024



Abstract:Since the emergence of the ImageNet dataset, the pretraining and fine-tuning approach has become widely adopted in computer vision due to the ability of ImageNet-pretrained models to learn a wide variety of visual features. However, a significant challenge arises when adapting these models to domain-specific fields, such as digital pathology, due to substantial gaps between domains. To address this limitation, foundation models (FM) have been trained on large-scale in-domain datasets to learn the intricate features of histopathology images. In cancer diagnosis, whole-slide image (WSI) prediction is essential for patient prognosis, and multiple instance learning (MIL) has been implemented to handle the giga-pixel size of WSI. As MIL frameworks rely on patch-level feature aggregation, this work aims to compare the performance of various feature extractors developed under different pretraining strategies for cancer subtyping on WSI under a MIL framework. Results demonstrate the ability of foundation models to surpass ImageNet-pretrained models for the prediction of six skin cancer subtypes
Self-Contrastive Weakly Supervised Learning Framework for Prognostic Prediction Using Whole Slide Images
May 24, 2024



Abstract:We present a pioneering investigation into the application of deep learning techniques to analyze histopathological images for addressing the substantial challenge of automated prognostic prediction. Prognostic prediction poses a unique challenge as the ground truth labels are inherently weak, and the model must anticipate future events that are not directly observable in the image. To address this challenge, we propose a novel three-part framework comprising of a convolutional network based tissue segmentation algorithm for region of interest delineation, a contrastive learning module for feature extraction, and a nested multiple instance learning classification module. Our study explores the significance of various regions of interest within the histopathological slides and exploits diverse learning scenarios. The pipeline is initially validated on artificially generated data and a simpler diagnostic task. Transitioning to prognostic prediction, tasks become more challenging. Employing bladder cancer as use case, our best models yield an AUC of 0.721 and 0.678 for recurrence and treatment outcome prediction respectively.
Speech emotion recognition from voice messages recorded in the wild
Mar 04, 2024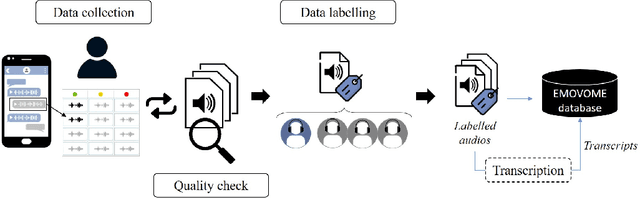

Abstract:Emotion datasets used for Speech Emotion Recognition (SER) often contain acted or elicited speech, limiting their applicability in real-world scenarios. In this work, we used the Emotional Voice Messages (EMOVOME) database, including spontaneous voice messages from conversations of 100 Spanish speakers on a messaging app, labeled in continuous and discrete emotions by expert and non-expert annotators. We created speaker-independent SER models using the eGeMAPS features, transformer-based models and their combination. We compared the results with reference databases and analyzed the influence of annotators and gender fairness. The pre-trained Unispeech-L model and its combination with eGeMAPS achieved the highest results, with 61.64% and 55.57% Unweighted Accuracy (UA) for 3-class valence and arousal prediction respectively, a 10% improvement over baseline models. For the emotion categories, 42.58% UA was obtained. EMOVOME performed lower than the acted RAVDESS database. The elicited IEMOCAP database also outperformed EMOVOME in the prediction of emotion categories, while similar results were obtained in valence and arousal. Additionally, EMOVOME outcomes varied with annotator labels, showing superior results and better fairness when combining expert and non-expert annotations. This study significantly contributes to the evaluation of SER models in real-life situations, advancing in the development of applications for analyzing spontaneous voice messages.
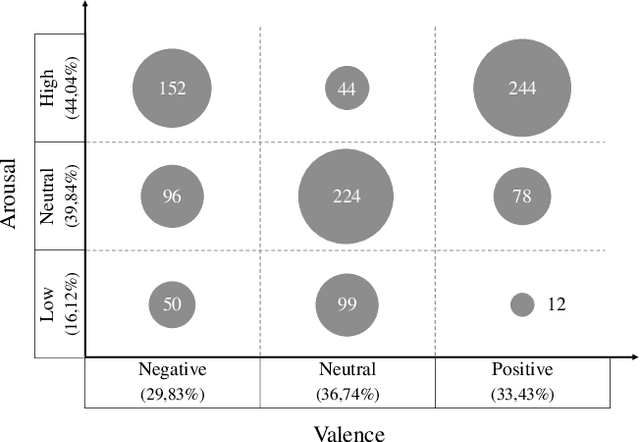
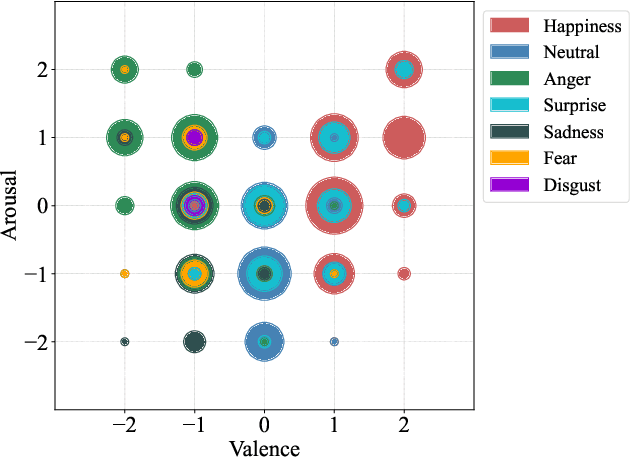
Emotional Voice Messages (EMOVOME) database: emotion recognition in spontaneous voice messages
Feb 27, 2024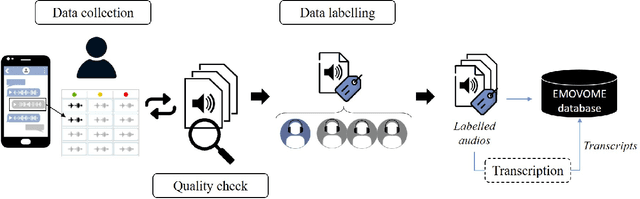



Abstract:Emotional Voice Messages (EMOVOME) is a spontaneous speech dataset containing 999 audio messages from real conversations on a messaging app from 100 Spanish speakers, gender balanced. Voice messages were produced in-the-wild conditions before participants were recruited, avoiding any conscious bias due to laboratory environment. Audios were labeled in valence and arousal dimensions by three non-experts and two experts, which were then combined to obtain a final label per dimension. The experts also provided an extra label corresponding to seven emotion categories. To set a baseline for future investigations using EMOVOME, we implemented emotion recognition models using both speech and audio transcriptions. For speech, we used the standard eGeMAPS feature set and support vector machines, obtaining 49.27% and 44.71% unweighted accuracy for valence and arousal respectively. For text, we fine-tuned a multilingual BERT model and achieved 61.15% and 47.43% unweighted accuracy for valence and arousal respectively. This database will significantly contribute to research on emotion recognition in the wild, while also providing a unique natural and freely accessible resource for Spanish.
Siamese Content-based Search Engine for a More Transparent Skin and Breast Cancer Diagnosis through Histological Imaging
Jan 16, 2024



Abstract:Computer Aid Diagnosis (CAD) has developed digital pathology with Deep Learning (DL)-based tools to assist pathologists in decision-making. Content-Based Histopathological Image Retrieval (CBHIR) is a novel tool to seek highly correlated patches in terms of similarity in histopathological features. In this work, we proposed two CBHIR approaches on breast (Breast-twins) and skin cancer (Skin-twins) data sets for robust and accurate patch-level retrieval, integrating a custom-built Siamese network as a feature extractor. The proposed Siamese network is able to generalize for unseen images by focusing on the similar histopathological features of the input pairs. The proposed CBHIR approaches are evaluated on the Breast (public) and Skin (private) data sets with top K accuracy. Finding the optimum amount of K is challenging, but also, as much as K increases, the dissimilarity between the query and the returned images increases which might mislead the pathologists. To the best of the author's belief, this paper is tackling this issue for the first time on histopathological images by evaluating the top first retrieved images. The Breast-twins model achieves 70% of the F1score at the top first, which exceeds the other state-of-the-art methods at a higher amount of K such as 5 and 400. Skin-twins overpasses the recently proposed Convolutional Auto Encoder (CAE) by 67%, increasing the precision. Besides, the Skin-twins model tackles the challenges of Spitzoid Tumors of Uncertain Malignant Potential (STUMP) to assist pathologists with retrieving top K images and their corresponding labels. So, this approach can offer a more explainable CAD tool to pathologists in terms of transparency, trustworthiness, or reliability among other characteristics.
Attention to detail: inter-resolution knowledge distillation
Jan 11, 2024Abstract:The development of computer vision solutions for gigapixel images in digital pathology is hampered by significant computational limitations due to the large size of whole slide images. In particular, digitizing biopsies at high resolutions is a time-consuming process, which is necessary due to the worsening results from the decrease in image detail. To alleviate this issue, recent literature has proposed using knowledge distillation to enhance the model performance at reduced image resolutions. In particular, soft labels and features extracted at the highest magnification level are distilled into a model that takes lower-magnification images as input. However, this approach fails to transfer knowledge about the most discriminative image regions in the classification process, which may be lost when the resolution is decreased. In this work, we propose to distill this information by incorporating attention maps during training. In particular, our formulation leverages saliency maps of the target class via grad-CAMs, which guides the lower-resolution Student model to match the Teacher distribution by minimizing the l2 distance between them. Comprehensive experiments on prostate histology image grading demonstrate that the proposed approach substantially improves the model performance across different image resolutions compared to previous literature.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge