Adrián Colomer
Defect Segmentation in OCT scans of ceramic parts for non-destructive inspection using deep learning
Oct 01, 2025



Abstract:Non-destructive testing (NDT) is essential in ceramic manufacturing to ensure the quality of components without compromising their integrity. In this context, Optical Coherence Tomography (OCT) enables high-resolution internal imaging, revealing defects such as pores, delaminations, or inclusions. This paper presents an automatic defect detection system based on Deep Learning (DL), trained on OCT images with manually segmented annotations. A neural network based on the U-Net architecture is developed, evaluating multiple experimental configurations to enhance its performance. Post-processing techniques enable both quantitative and qualitative evaluation of the predictions. The system shows an accurate behavior of 0.979 Dice Score, outperforming comparable studies. The inference time of 18.98 seconds per volume supports its viability for detecting inclusions, enabling more efficient, reliable, and automated quality control.
Siamese Content-based Search Engine for a More Transparent Skin and Breast Cancer Diagnosis through Histological Imaging
Jan 16, 2024



Abstract:Computer Aid Diagnosis (CAD) has developed digital pathology with Deep Learning (DL)-based tools to assist pathologists in decision-making. Content-Based Histopathological Image Retrieval (CBHIR) is a novel tool to seek highly correlated patches in terms of similarity in histopathological features. In this work, we proposed two CBHIR approaches on breast (Breast-twins) and skin cancer (Skin-twins) data sets for robust and accurate patch-level retrieval, integrating a custom-built Siamese network as a feature extractor. The proposed Siamese network is able to generalize for unseen images by focusing on the similar histopathological features of the input pairs. The proposed CBHIR approaches are evaluated on the Breast (public) and Skin (private) data sets with top K accuracy. Finding the optimum amount of K is challenging, but also, as much as K increases, the dissimilarity between the query and the returned images increases which might mislead the pathologists. To the best of the author's belief, this paper is tackling this issue for the first time on histopathological images by evaluating the top first retrieved images. The Breast-twins model achieves 70% of the F1score at the top first, which exceeds the other state-of-the-art methods at a higher amount of K such as 5 and 400. Skin-twins overpasses the recently proposed Convolutional Auto Encoder (CAE) by 67%, increasing the precision. Besides, the Skin-twins model tackles the challenges of Spitzoid Tumors of Uncertain Malignant Potential (STUMP) to assist pathologists with retrieving top K images and their corresponding labels. So, this approach can offer a more explainable CAD tool to pathologists in terms of transparency, trustworthiness, or reliability among other characteristics.
Attention to detail: inter-resolution knowledge distillation
Jan 11, 2024Abstract:The development of computer vision solutions for gigapixel images in digital pathology is hampered by significant computational limitations due to the large size of whole slide images. In particular, digitizing biopsies at high resolutions is a time-consuming process, which is necessary due to the worsening results from the decrease in image detail. To alleviate this issue, recent literature has proposed using knowledge distillation to enhance the model performance at reduced image resolutions. In particular, soft labels and features extracted at the highest magnification level are distilled into a model that takes lower-magnification images as input. However, this approach fails to transfer knowledge about the most discriminative image regions in the classification process, which may be lost when the resolution is decreased. In this work, we propose to distill this information by incorporating attention maps during training. In particular, our formulation leverages saliency maps of the target class via grad-CAMs, which guides the lower-resolution Student model to match the Teacher distribution by minimizing the l2 distance between them. Comprehensive experiments on prostate histology image grading demonstrate that the proposed approach substantially improves the model performance across different image resolutions compared to previous literature.
WWFedCBMIR: World-Wide Federated Content-Based Medical Image Retrieval
May 05, 2023Abstract:The paper proposes a Federated Content-Based Medical Image Retrieval (FedCBMIR) platform that utilizes Federated Learning (FL) to address the challenges of acquiring a diverse medical data set for training CBMIR models. CBMIR assists pathologists in diagnosing breast cancer more rapidly by identifying similar medical images and relevant patches in prior cases compared to traditional cancer detection methods. However, CBMIR in histopathology necessitates a pool of Whole Slide Images (WSIs) to train to extract an optimal embedding vector that leverages search engine performance, which may not be available in all centers. The strict regulations surrounding data sharing in medical data sets also hinder research and model development, making it difficult to collect a rich data set. The proposed FedCBMIR distributes the model to collaborative centers for training without sharing the data set, resulting in shorter training times than local training. FedCBMIR was evaluated in two experiments with three scenarios on BreaKHis and Camelyon17 (CAM17). The study shows that the FedCBMIR method increases the F1-Score (F1S) of each client to 98%, 96%, 94%, and 97% in the BreaKHis experiment with a generalized model of four magnifications and does so in 6.30 hours less time than total local training. FedCBMIR also achieves 98% accuracy with CAM17 in 2.49 hours less training time than local training, demonstrating that our FedCBMIR is both fast and accurate for both pathologists and engineers. In addition, our FedCBMIR provides similar images with higher magnification for non-developed countries where participate in the worldwide FedCBMIR with developed countries to facilitate mitosis measuring in breast cancer diagnosis. We evaluate this scenario by scattering BreaKHis into four centers with different magnifications.
A self-training framework for glaucoma grading in OCT B-scans
Nov 23, 2021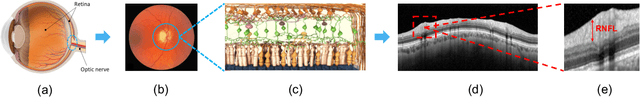
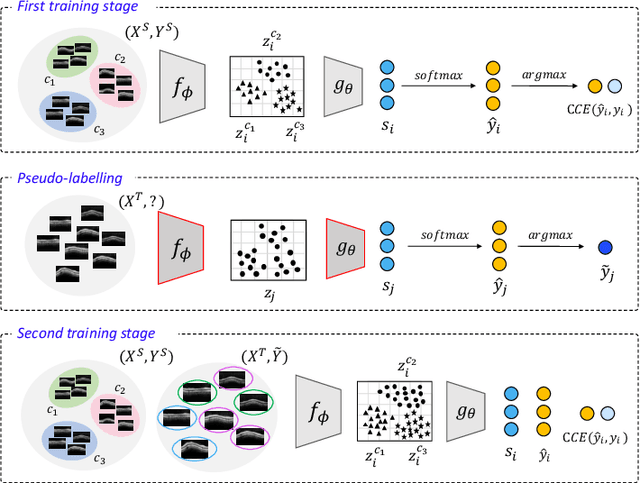
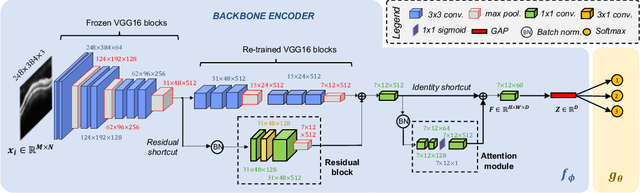
Abstract:In this paper, we present a self-training-based framework for glaucoma grading using OCT B-scans under the presence of domain shift. Particularly, the proposed two-step learning methodology resorts to pseudo-labels generated during the first step to augment the training dataset on the target domain, which is then used to train the final target model. This allows transferring knowledge-domain from the unlabeled data. Additionally, we propose a novel glaucoma-specific backbone which introduces residual and attention modules via skip-connections to refine the embedding features of the latent space. By doing this, our model is capable of improving state-of-the-art from a quantitative and interpretability perspective. The reported results demonstrate that the proposed learning strategy can boost the performance of the model on the target dataset without incurring in additional annotation steps, by using only labels from the source examples. Our model consistently outperforms the baseline by 1-3% across different metrics and bridges the gap with respect to training the model on the labeled target data.
A Novel Self-Learning Framework for Bladder Cancer Grading Using Histopathological Images
Jun 25, 2021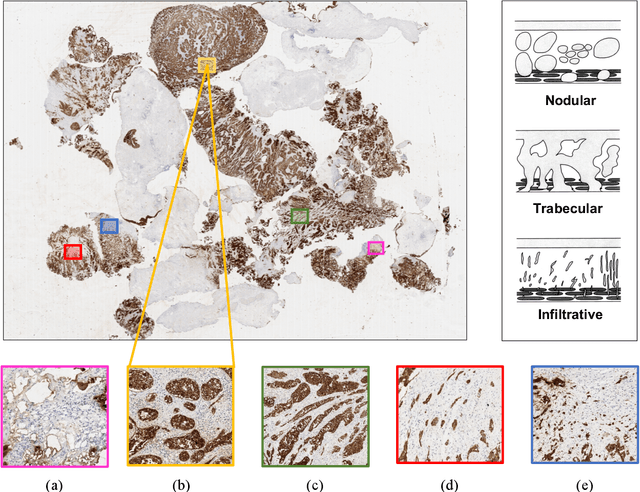

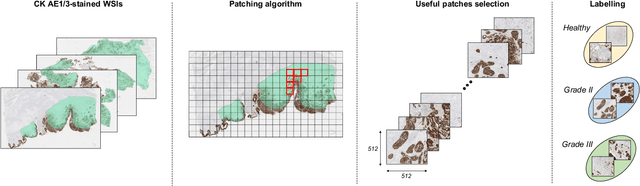

Abstract:Recently, bladder cancer has been significantly increased in terms of incidence and mortality. Currently, two subtypes are known based on tumour growth: non-muscle invasive (NMIBC) and muscle-invasive bladder cancer (MIBC). In this work, we focus on the MIBC subtype because it is of the worst prognosis and can spread to adjacent organs. We present a self-learning framework to grade bladder cancer from histological images stained via immunohistochemical techniques. Specifically, we propose a novel Deep Convolutional Embedded Attention Clustering (DCEAC) which allows classifying histological patches into different severity levels of the disease, according to the patterns established in the literature. The proposed DCEAC model follows a two-step fully unsupervised learning methodology to discern between non-tumour, mild and infiltrative patterns from high-resolution samples of 512x512 pixels. Our system outperforms previous clustering-based methods by including a convolutional attention module, which allows refining the features of the latent space before the classification stage. The proposed network exceeds state-of-the-art approaches by 2-3% across different metrics, achieving a final average accuracy of 0.9034 in a multi-class scenario. Furthermore, the reported class activation maps evidence that our model is able to learn by itself the same patterns that clinicians consider relevant, without incurring prior annotation steps. This fact supposes a breakthrough in muscle-invasive bladder cancer grading which bridges the gap with respect to train the model on labelled data.
Circumpapillary OCT-Focused Hybrid Learning for Glaucoma Grading Using Tailored Prototypical Neural Networks
Jun 25, 2021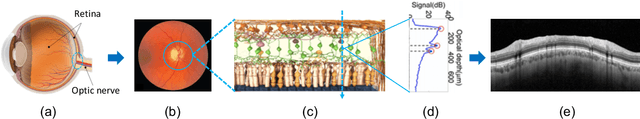

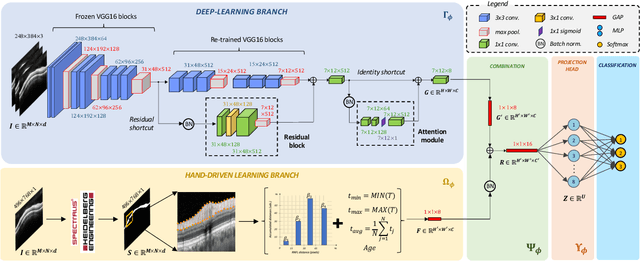

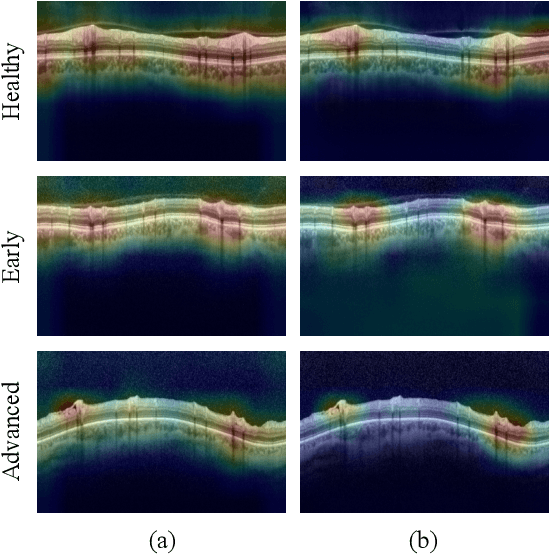
Abstract:Glaucoma is one of the leading causes of blindness worldwide and Optical Coherence Tomography (OCT) is the quintessential imaging technique for its detection. Unlike most of the state-of-the-art studies focused on glaucoma detection, in this paper, we propose, for the first time, a novel framework for glaucoma grading using raw circumpapillary B-scans. In particular, we set out a new OCT-based hybrid network which combines hand-driven and deep learning algorithms. An OCT-specific descriptor is proposed to extract hand-crafted features related to the retinal nerve fibre layer (RNFL). In parallel, an innovative CNN is developed using skip-connections to include tailored residual and attention modules to refine the automatic features of the latent space. The proposed architecture is used as a backbone to conduct a novel few-shot learning based on static and dynamic prototypical networks. The k-shot paradigm is redefined giving rise to a supervised end-to-end system which provides substantial improvements discriminating between healthy, early and advanced glaucoma samples. The training and evaluation processes of the dynamic prototypical network are addressed from two fused databases acquired via Heidelberg Spectralis system. Validation and testing results reach a categorical accuracy of 0.9459 and 0.8788 for glaucoma grading, respectively. Besides, the high performance reported by the proposed model for glaucoma detection deserves a special mention. The findings from the class activation maps are directly in line with the clinicians' opinion since the heatmaps pointed out the RNFL as the most relevant structure for glaucoma diagnosis.
Prostate Gland Segmentation in Histology Images via Residual and Multi-Resolution U-Net
May 21, 2021
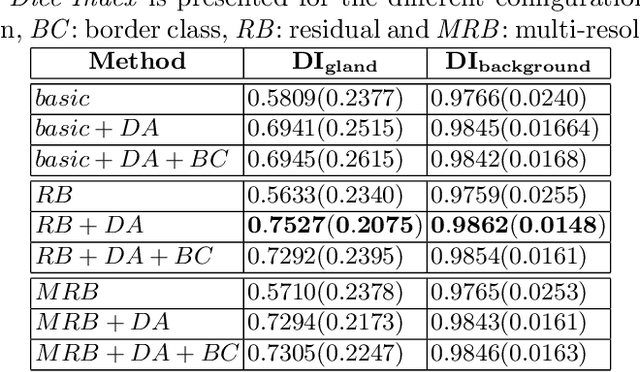
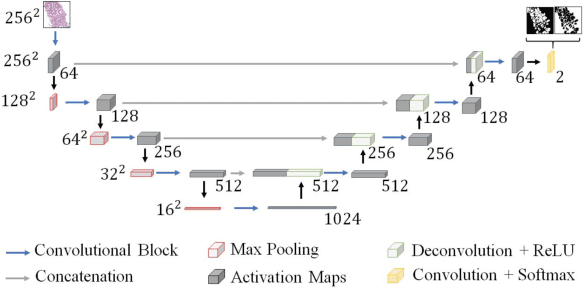
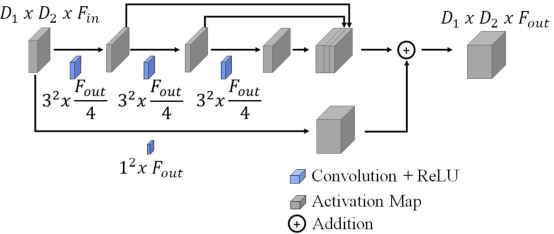
Abstract:Prostate cancer is one of the most prevalent cancers worldwide. One of the key factors in reducing its mortality is based on early detection. The computer-aided diagnosis systems for this task are based on the glandular structural analysis in histology images. Hence, accurate gland detection and segmentation is crucial for a successful prediction. The methodological basis of this work is a prostate gland segmentation based on U-Net convolutional neural network architectures modified with residual and multi-resolution blocks, trained using data augmentation techniques. The residual configuration outperforms in the test subset the previous state-of-the-art approaches in an image-level comparison, reaching an average Dice Index of 0.77.
Going Deeper through the Gleason Scoring Scale: An Automatic end-to-end System for Histology Prostate Grading and Cribriform Pattern Detection
May 21, 2021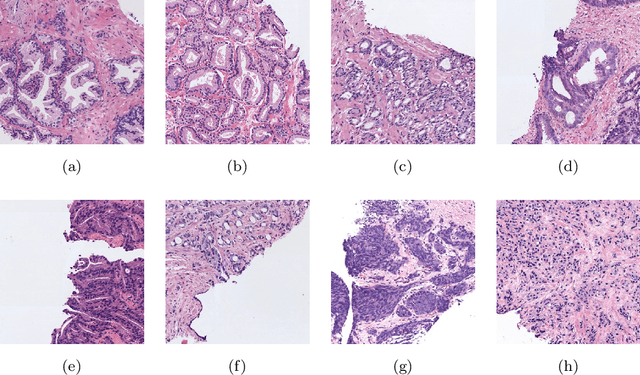



Abstract:The Gleason scoring system is the primary diagnostic and prognostic tool for prostate cancer. In recent years, with the development of digitisation devices, the use of computer vision techniques for the analysis of biopsies has increased. However, to the best of the authors' knowledge, the development of algorithms to automatically detect individual cribriform patterns belonging to Gleason grade 4 has not yet been studied in the literature. The objective of the work presented in this paper is to develop a deep-learning-based system able to support pathologists in the daily analysis of prostate biopsies. The methodological core of this work is a patch-wise predictive model based on convolutional neural networks able to determine the presence of cancerous patterns. In particular, we train from scratch a simple self-design architecture. The cribriform pattern is detected by retraining the set of filters of the last convolutional layer in the network. From the reconstructed prediction map, we compute the percentage of each Gleason grade in the tissue to feed a multi-layer perceptron which provides a biopsy-level score.mIn our SICAPv2 database, composed of 182 annotated whole slide images, we obtained a Cohen's quadratic kappa of 0.77 in the test set for the patch-level Gleason grading with the proposed architecture trained from scratch. Our results outperform previous ones reported in the literature. Furthermore, this model reaches the level of fine-tuned state-of-the-art architectures in a patient-based four groups cross validation. In the cribriform pattern detection task, we obtained an area under ROC curve of 0.82. Regarding the biopsy Gleason scoring, we achieved a quadratic Cohen's Kappa of 0.81 in the test subset. Shallow CNN architectures trained from scratch outperform current state-of-the-art methods for Gleason grades classification.
WeGleNet: A Weakly-Supervised Convolutional Neural Network for the Semantic Segmentation of Gleason Grades in Prostate Histology Images
May 21, 2021



Abstract:Prostate cancer is one of the main diseases affecting men worldwide. The Gleason scoring system is the primary diagnostic tool for prostate cancer. This is obtained via the visual analysis of cancerous patterns in prostate biopsies performed by expert pathologists, and the aggregation of the main Gleason grades in a combined score. Computer-aided diagnosis systems allow to reduce the workload of pathologists and increase the objectivity. Recently, efforts have been made in the literature to develop algorithms aiming the direct estimation of the global Gleason score at biopsy/core level with global labels. However, these algorithms do not cover the accurate localization of the Gleason patterns into the tissue. In this work, we propose a deep-learning-based system able to detect local cancerous patterns in the prostate tissue using only the global-level Gleason score during training. The methodological core of this work is the proposed weakly-supervised-trained convolutional neural network, WeGleNet, based on a multi-class segmentation layer after the feature extraction module, a global-aggregation, and the slicing of the background class for the model loss estimation during training. We obtained a Cohen's quadratic kappa (k) of 0.67 for the pixel-level prediction of cancerous patterns in the validation cohort. We compared the model performance for semantic segmentation of Gleason grades with supervised state-of-the-art architectures in the test cohort. We obtained a pixel-level k of 0.61 and a macro-averaged f1-score of 0.58, at the same level as fully-supervised methods. Regarding the estimation of the core-level Gleason score, we obtained a k of 0.76 and 0.67 between the model and two different pathologists. WeGleNet is capable of performing the semantic segmentation of Gleason grades similarly to fully-supervised methods without requiring pixel-level annotations.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge