Yuandou Wang
IMLP: An Energy-Efficient Continual Learning Method for Tabular Data Streams
Oct 06, 2025Abstract:Tabular data streams are rapidly emerging as a dominant modality for real-time decision-making in healthcare, finance, and the Internet of Things (IoT). These applications commonly run on edge and mobile devices, where energy budgets, memory, and compute are strictly limited. Continual learning (CL) addresses such dynamics by training models sequentially on task streams while preserving prior knowledge and consolidating new knowledge. While recent CL work has advanced in mitigating catastrophic forgetting and improving knowledge transfer, the practical requirements of energy and memory efficiency for tabular data streams remain underexplored. In particular, existing CL solutions mostly depend on replay mechanisms whose buffers grow over time and exacerbate resource costs. We propose a context-aware incremental Multi-Layer Perceptron (IMLP), a compact continual learner for tabular data streams. IMLP incorporates a windowed scaled dot-product attention over a sliding latent feature buffer, enabling constant-size memory and avoiding storing raw data. The attended context is concatenated with current features and processed by shared feed-forward layers, yielding lightweight per-segment updates. To assess practical deployability, we introduce NetScore-T, a tunable metric coupling balanced accuracy with energy for Pareto-aware comparison across models and datasets. IMLP achieves up to $27.6\times$ higher energy efficiency than TabNet and $85.5\times$ higher than TabPFN, while maintaining competitive average accuracy. Overall, IMLP provides an easy-to-deploy, energy-efficient alternative to full retraining for tabular data streams.
PriCE: Privacy-Preserving and Cost-Effective Scheduling for Parallelizing the Large Medical Image Processing Workflow over Hybrid Clouds
May 24, 2024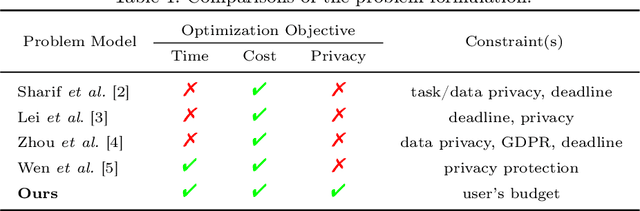


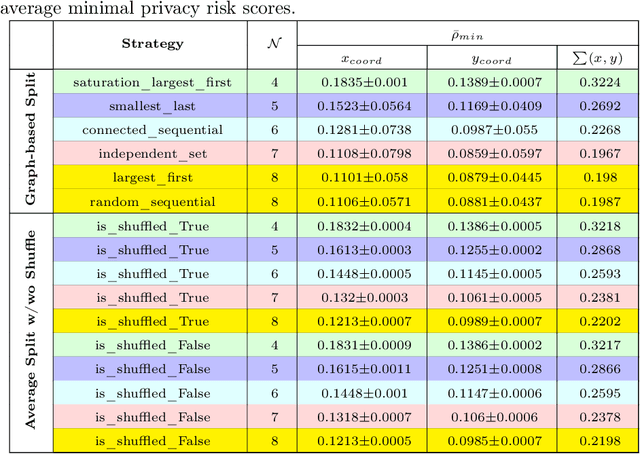
Abstract:Running deep neural networks for large medical images is a resource-hungry and time-consuming task with centralized computing. Outsourcing such medical image processing tasks to hybrid clouds has benefits, such as a significant reduction of execution time and monetary cost. However, due to privacy concerns, it is still challenging to process sensitive medical images over clouds, which would hinder their deployment in many real-world applications. To overcome this, we first formulate the overall optimization objectives of the privacy-preserving distributed system model, i.e., minimizing the amount of information about the private data learned by the adversaries throughout the process, reducing the maximum execution time and cost under the user budget constraint. We propose a novel privacy-preserving and cost-effective method called PriCE to solve this multi-objective optimization problem. We performed extensive simulation experiments for artifact detection tasks on medical images using an ensemble of five deep convolutional neural network inferences as the workflow task. Experimental results show that PriCE successfully splits a wide range of input gigapixel medical images with graph-coloring-based strategies, yielding desired output utility and lowering the privacy risk, makespan, and monetary cost under user's budget.
WWFedCBMIR: World-Wide Federated Content-Based Medical Image Retrieval
May 05, 2023Abstract:The paper proposes a Federated Content-Based Medical Image Retrieval (FedCBMIR) platform that utilizes Federated Learning (FL) to address the challenges of acquiring a diverse medical data set for training CBMIR models. CBMIR assists pathologists in diagnosing breast cancer more rapidly by identifying similar medical images and relevant patches in prior cases compared to traditional cancer detection methods. However, CBMIR in histopathology necessitates a pool of Whole Slide Images (WSIs) to train to extract an optimal embedding vector that leverages search engine performance, which may not be available in all centers. The strict regulations surrounding data sharing in medical data sets also hinder research and model development, making it difficult to collect a rich data set. The proposed FedCBMIR distributes the model to collaborative centers for training without sharing the data set, resulting in shorter training times than local training. FedCBMIR was evaluated in two experiments with three scenarios on BreaKHis and Camelyon17 (CAM17). The study shows that the FedCBMIR method increases the F1-Score (F1S) of each client to 98%, 96%, 94%, and 97% in the BreaKHis experiment with a generalized model of four magnifications and does so in 6.30 hours less time than total local training. FedCBMIR also achieves 98% accuracy with CAM17 in 2.49 hours less training time than local training, demonstrating that our FedCBMIR is both fast and accurate for both pathologists and engineers. In addition, our FedCBMIR provides similar images with higher magnification for non-developed countries where participate in the worldwide FedCBMIR with developed countries to facilitate mitosis measuring in breast cancer diagnosis. We evaluate this scenario by scattering BreaKHis into four centers with different magnifications.
A Survey on Dataset Distillation: Approaches, Applications and Future Directions
May 03, 2023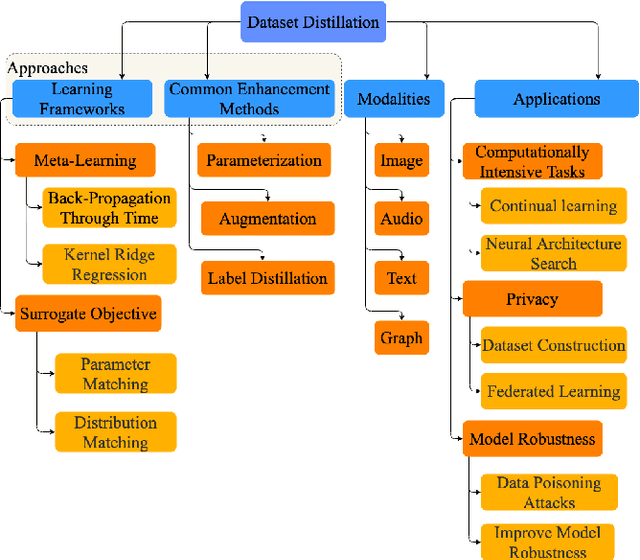
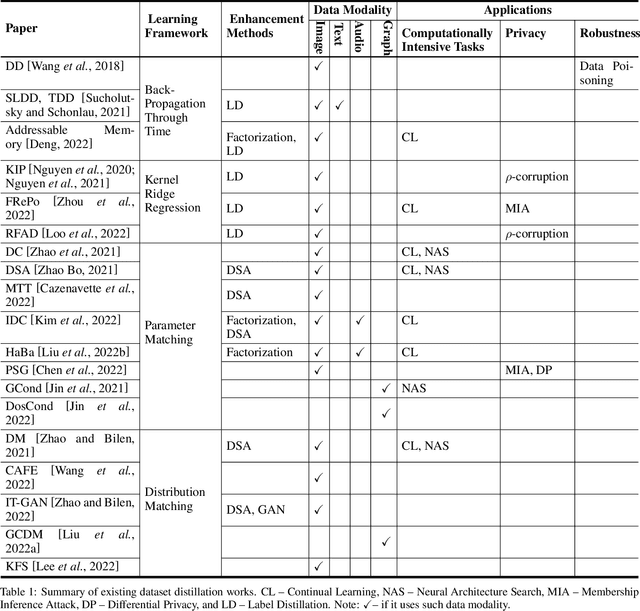
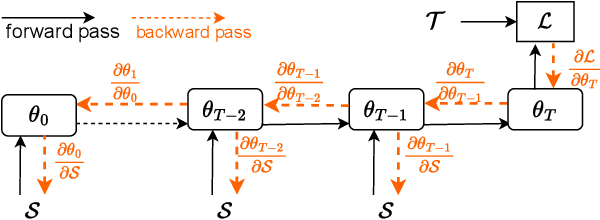

Abstract:Dataset distillation is attracting more attention in machine learning as training sets continue to grow and the cost of training state-of-the-art models becomes increasingly high. By synthesizing datasets with high information density, dataset distillation offers a range of potential applications, including support for continual learning, neural architecture search, and privacy protection. Despite recent advances, we lack a holistic understanding of the approaches and applications. Our survey aims to bridge this gap by first proposing a taxonomy of dataset distillation, characterizing existing approaches, and then systematically reviewing the data modalities, and related applications. In addition, we summarize the challenges and discuss future directions for this field of research.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge