Kjersti Engan
Two-Stream Thermal Imaging Fusion for Enhanced Time of Birth Detection in Neonatal Care
Mar 05, 2025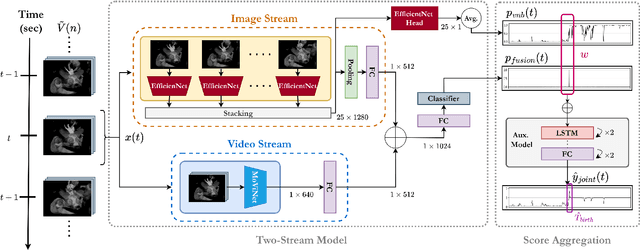
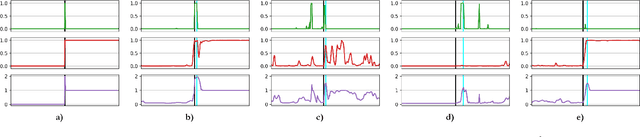
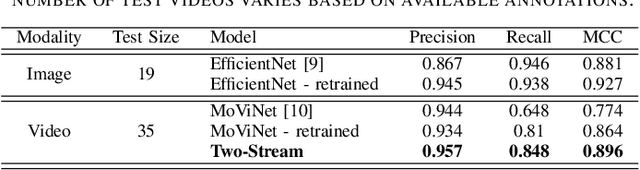
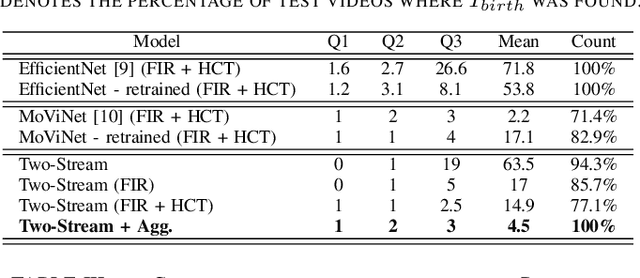
Abstract:Around 10% of newborns require some help to initiate breathing, and 5\% need ventilation assistance. Accurate Time of Birth (ToB) documentation is essential for optimizing neonatal care, as timely interventions are vital for proper resuscitation. However, current clinical methods for recording ToB often rely on manual processes, which can be prone to inaccuracies. In this study, we present a novel two-stream fusion system that combines the power of image and video analysis to accurately detect the ToB from thermal recordings in the delivery room and operating theater. By integrating static and dynamic streams, our approach captures richer birth-related spatiotemporal features, leading to more robust and precise ToB estimation. We demonstrate that this synergy between data modalities enhances performance over single-stream approaches. Our system achieves 95.7% precision and 84.8% recall in detecting birth within short video clips. Additionally, with the help of a score aggregation module, it successfully identifies ToB in 100% of test cases, with a median absolute error of 2 seconds and an absolute mean deviation of 4.5 seconds compared to manual annotations.
AI-Based Thermal Video Analysis in Privacy-Preserving Healthcare: A Case Study on Detecting Time of Birth
Feb 05, 2025Abstract:Approximately 10% of newborns need some assistance to start breathing and 5\% proper ventilation. It is crucial that interventions are initiated as soon as possible after birth. Accurate documentation of Time of Birth (ToB) is thereby essential for documenting and improving newborn resuscitation performance. However, current clinical practices rely on manual recording of ToB, typically with minute precision. In this study, we present an AI-driven, video-based system for automated ToB detection using thermal imaging, designed to preserve the privacy of healthcare providers and mothers by avoiding the use of identifiable visual data. Our approach achieves 91.4% precision and 97.4% recall in detecting ToB within thermal video clips during performance evaluation. Additionally, our system successfully identifies ToB in 96% of test cases with an absolute median deviation of 1 second compared to manual annotations. This method offers a reliable solution for improving ToB documentation and enhancing newborn resuscitation outcomes.
Advancing Newborn Care: Precise Birth Time Detection Using AI-Driven Thermal Imaging with Adaptive Normalization
Oct 14, 2024Abstract:Around 5-10\% of newborns need assistance to start breathing. Currently, there is a lack of evidence-based research, objective data collection, and opportunities for learning from real newborn resuscitation emergency events. Generating and evaluating automated newborn resuscitation algorithm activity timelines relative to the Time of Birth (ToB) offers a promising opportunity to enhance newborn care practices. Given the importance of prompt resuscitation interventions within the "golden minute" after birth, having an accurate ToB with second precision is essential for effective subsequent analysis of newborn resuscitation episodes. Instead, ToB is generally registered manually, often with minute precision, making the process inefficient and susceptible to error and imprecision. In this work, we explore the fusion of Artificial Intelligence (AI) and thermal imaging to develop the first AI-driven ToB detector. The use of temperature information offers a promising alternative to detect the newborn while respecting the privacy of healthcare providers and mothers. However, the frequent inconsistencies in thermal measurements, especially in a multi-camera setup, make normalization strategies critical. Our methodology involves a three-step process: first, we propose an adaptive normalization method based on Gaussian mixture models (GMM) to mitigate issues related to temperature variations; second, we implement and deploy an AI model to detect the presence of the newborn within the thermal video frames; and third, we evaluate and post-process the model's predictions to estimate the ToB. A precision of 88.1\% and a recall of 89.3\% are reported in the detection of the newborn within thermal frames during performance evaluation. Our approach achieves an absolute median deviation of 2.7 seconds in estimating the ToB relative to the manual annotations.
NMGrad: Advancing Histopathological Bladder Cancer Grading with Weakly Supervised Deep Learning
May 24, 2024Abstract:The most prevalent form of bladder cancer is urothelial carcinoma, characterized by a high recurrence rate and substantial lifetime treatment costs for patients. Grading is a prime factor for patient risk stratification, although it suffers from inconsistencies and variations among pathologists. Moreover, absence of annotations in medical imaging difficults training deep learning models. To address these challenges, we introduce a pipeline designed for bladder cancer grading using histological slides. First, it extracts urothelium tissue tiles at different magnification levels, employing a convolutional neural network for processing for feature extraction. Then, it engages in the slide-level prediction process. It employs a nested multiple instance learning approach with attention to predict the grade. To distinguish different levels of malignancy within specific regions of the slide, we include the origins of the tiles in our analysis. The attention scores at region level is shown to correlate with verified high-grade regions, giving some explainability to the model. Clinical evaluations demonstrate that our model consistently outperforms previous state-of-the-art methods.
PriCE: Privacy-Preserving and Cost-Effective Scheduling for Parallelizing the Large Medical Image Processing Workflow over Hybrid Clouds
May 24, 2024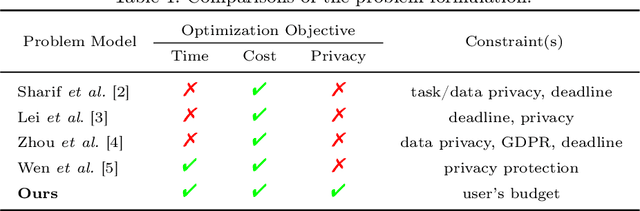


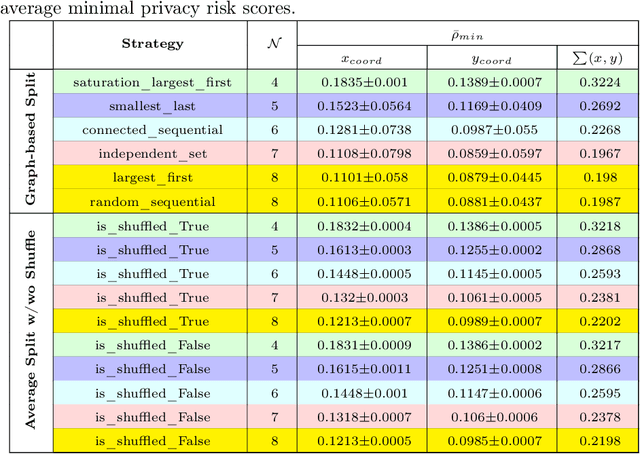
Abstract:Running deep neural networks for large medical images is a resource-hungry and time-consuming task with centralized computing. Outsourcing such medical image processing tasks to hybrid clouds has benefits, such as a significant reduction of execution time and monetary cost. However, due to privacy concerns, it is still challenging to process sensitive medical images over clouds, which would hinder their deployment in many real-world applications. To overcome this, we first formulate the overall optimization objectives of the privacy-preserving distributed system model, i.e., minimizing the amount of information about the private data learned by the adversaries throughout the process, reducing the maximum execution time and cost under the user budget constraint. We propose a novel privacy-preserving and cost-effective method called PriCE to solve this multi-objective optimization problem. We performed extensive simulation experiments for artifact detection tasks on medical images using an ensemble of five deep convolutional neural network inferences as the workflow task. Experimental results show that PriCE successfully splits a wide range of input gigapixel medical images with graph-coloring-based strategies, yielding desired output utility and lowering the privacy risk, makespan, and monetary cost under user's budget.
Self-Contrastive Weakly Supervised Learning Framework for Prognostic Prediction Using Whole Slide Images
May 24, 2024



Abstract:We present a pioneering investigation into the application of deep learning techniques to analyze histopathological images for addressing the substantial challenge of automated prognostic prediction. Prognostic prediction poses a unique challenge as the ground truth labels are inherently weak, and the model must anticipate future events that are not directly observable in the image. To address this challenge, we propose a novel three-part framework comprising of a convolutional network based tissue segmentation algorithm for region of interest delineation, a contrastive learning module for feature extraction, and a nested multiple instance learning classification module. Our study explores the significance of various regions of interest within the histopathological slides and exploits diverse learning scenarios. The pipeline is initially validated on artificially generated data and a simpler diagnostic task. Transitioning to prognostic prediction, tasks become more challenging. Employing bladder cancer as use case, our best models yield an AUC of 0.721 and 0.678 for recurrence and treatment outcome prediction respectively.
Equipping Computational Pathology Systems with Artifact Processing Pipelines: A Showcase for Computation and Performance Trade-offs
Mar 13, 2024Abstract:Histopathology is a gold standard for cancer diagnosis under a microscopic examination. However, histological tissue processing procedures result in artifacts, which are ultimately transferred to the digitized version of glass slides, known as whole slide images (WSIs). Artifacts are diagnostically irrelevant areas and may result in wrong deep learning (DL) algorithms predictions. Therefore, detecting and excluding artifacts in the computational pathology (CPATH) system is essential for reliable automated diagnosis. In this paper, we propose a mixture of experts (MoE) scheme for detecting five notable artifacts, including damaged tissue, blur, folded tissue, air bubbles, and histologically irrelevant blood from WSIs. First, we train independent binary DL models as experts to capture particular artifact morphology. Then, we ensemble their predictions using a fusion mechanism. We apply probabilistic thresholding over the final probability distribution to improve the sensitivity of the MoE. We developed DL pipelines using two MoEs and two multiclass models of state-of-the-art deep convolutional neural networks (DCNNs) and vision transformers (ViTs). DCNNs-based MoE and ViTs-based MoE schemes outperformed simpler multiclass models and were tested on datasets from different hospitals and cancer types, where MoE using DCNNs yielded the best results. The proposed MoE yields 86.15% F1 and 97.93% sensitivity scores on unseen data, retaining less computational cost for inference than MoE using ViTs. This best performance of MoEs comes with relatively higher computational trade-offs than multiclass models. The proposed artifact detection pipeline will not only ensure reliable CPATH predictions but may also provide quality control.
A Dual Convolutional Neural Network Pipeline for Melanoma Diagnostics and Prognostics
Dec 14, 2023Abstract:Melanoma is a type of cancer that begins in the cells controlling the pigment of the skin, and it is often referred to as the most dangerous skin cancer. Diagnosing melanoma can be time-consuming, and a recent increase in melanoma incidents indicates a growing demand for a more efficient diagnostic process. This paper presents a pipeline for melanoma diagnostics, leveraging two convolutional neural networks, a diagnosis, and a prognosis model. The diagnostic model is responsible for localizing malignant patches across whole slide images and delivering a patient-level diagnosis as malignant or benign. Further, the prognosis model utilizes the diagnostic model's output to provide a patient-level prognosis as good or bad. The full pipeline has an F1 score of 0.79 when tested on data from the same distribution as it was trained on.
Balancing Privacy and Progress in Artificial Intelligence: Anonymization in Histopathology for Biomedical Research and Education
Aug 08, 2023Abstract:The advancement of biomedical research heavily relies on access to large amounts of medical data. In the case of histopathology, Whole Slide Images (WSI) and clinicopathological information are valuable for developing Artificial Intelligence (AI) algorithms for Digital Pathology (DP). Transferring medical data "as open as possible" enhances the usability of the data for secondary purposes but poses a risk to patient privacy. At the same time, existing regulations push towards keeping medical data "as closed as necessary" to avoid re-identification risks. Generally, these legal regulations require the removal of sensitive data but do not consider the possibility of data linkage attacks due to modern image-matching algorithms. In addition, the lack of standardization in DP makes it harder to establish a single solution for all formats of WSIs. These challenges raise problems for bio-informatics researchers in balancing privacy and progress while developing AI algorithms. This paper explores the legal regulations and terminologies for medical data-sharing. We review existing approaches and highlight challenges from the histopathological perspective. We also present a data-sharing guideline for histological data to foster multidisciplinary research and education.
Vision Transformers for Small Histological Datasets Learned through Knowledge Distillation
May 27, 2023Abstract:Computational Pathology (CPATH) systems have the potential to automate diagnostic tasks. However, the artifacts on the digitized histological glass slides, known as Whole Slide Images (WSIs), may hamper the overall performance of CPATH systems. Deep Learning (DL) models such as Vision Transformers (ViTs) may detect and exclude artifacts before running the diagnostic algorithm. A simple way to develop robust and generalized ViTs is to train them on massive datasets. Unfortunately, acquiring large medical datasets is expensive and inconvenient, prompting the need for a generalized artifact detection method for WSIs. In this paper, we present a student-teacher recipe to improve the classification performance of ViT for the air bubbles detection task. ViT, trained under the student-teacher framework, boosts its performance by distilling existing knowledge from the high-capacity teacher model. Our best-performing ViT yields 0.961 and 0.911 F1-score and MCC, respectively, observing a 7% gain in MCC against stand-alone training. The proposed method presents a new perspective of leveraging knowledge distillation over transfer learning to encourage the use of customized transformers for efficient preprocessing pipelines in the CPATH systems.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge