Rafael Molina
torchmil: A PyTorch-based library for deep Multiple Instance Learning
Sep 09, 2025Abstract:Multiple Instance Learning (MIL) is a powerful framework for weakly supervised learning, particularly useful when fine-grained annotations are unavailable. Despite growing interest in deep MIL methods, the field lacks standardized tools for model development, evaluation, and comparison, which hinders reproducibility and accessibility. To address this, we present torchmil, an open-source Python library built on PyTorch. torchmil offers a unified, modular, and extensible framework, featuring basic building blocks for MIL models, a standardized data format, and a curated collection of benchmark datasets and models. The library includes comprehensive documentation and tutorials to support both practitioners and researchers. torchmil aims to accelerate progress in MIL and lower the entry barrier for new users. Available at https://torchmil.readthedocs.io.
Sm: enhanced localization in Multiple Instance Learning for medical imaging classification
Oct 04, 2024Abstract:Multiple Instance Learning (MIL) is widely used in medical imaging classification to reduce the labeling effort. While only bag labels are available for training, one typically seeks predictions at both bag and instance levels (classification and localization tasks, respectively). Early MIL methods treated the instances in a bag independently. Recent methods account for global and local dependencies among instances. Although they have yielded excellent results in classification, their performance in terms of localization is comparatively limited. We argue that these models have been designed to target the classification task, while implications at the instance level have not been deeply investigated. Motivated by a simple observation -- that neighboring instances are likely to have the same label -- we propose a novel, principled, and flexible mechanism to model local dependencies. It can be used alone or combined with any mechanism to model global dependencies (e.g., transformers). A thorough empirical validation shows that our module leads to state-of-the-art performance in localization while being competitive or superior in classification. Our code is at https://github.com/Franblueee/SmMIL.
Focused Active Learning for Histopathological Image Classification
Apr 06, 2024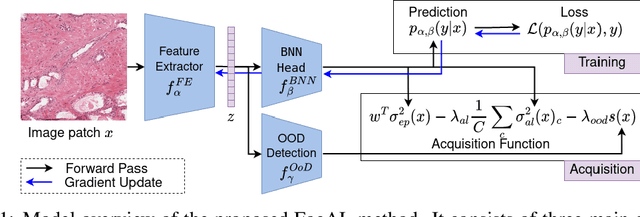
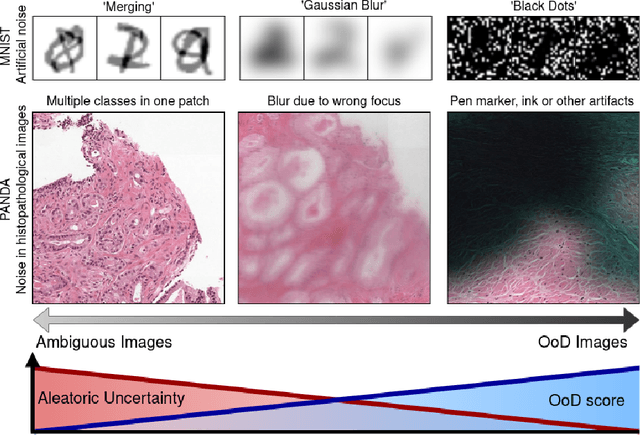
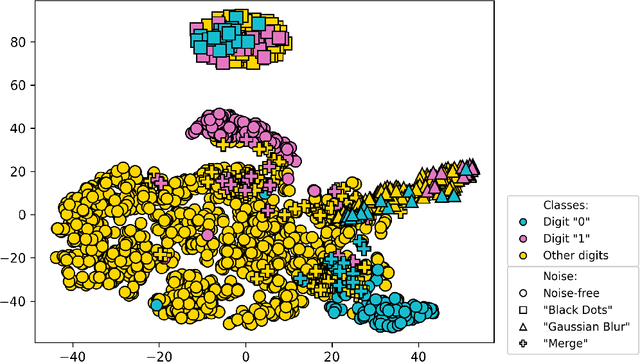
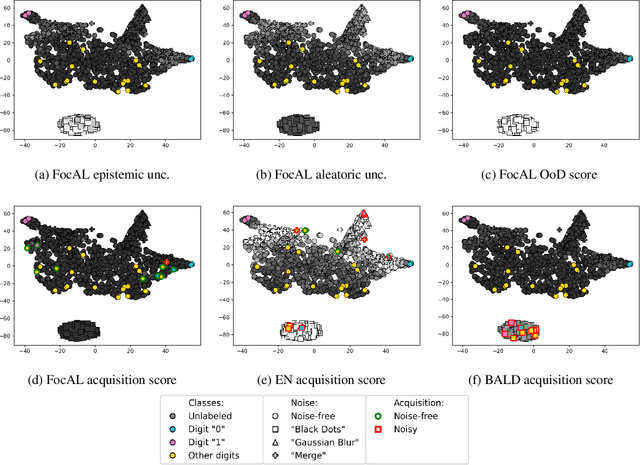
Abstract:Active Learning (AL) has the potential to solve a major problem of digital pathology: the efficient acquisition of labeled data for machine learning algorithms. However, existing AL methods often struggle in realistic settings with artifacts, ambiguities, and class imbalances, as commonly seen in the medical field. The lack of precise uncertainty estimations leads to the acquisition of images with a low informative value. To address these challenges, we propose Focused Active Learning (FocAL), which combines a Bayesian Neural Network with Out-of-Distribution detection to estimate different uncertainties for the acquisition function. Specifically, the weighted epistemic uncertainty accounts for the class imbalance, aleatoric uncertainty for ambiguous images, and an OoD score for artifacts. We perform extensive experiments to validate our method on MNIST and the real-world Panda dataset for the classification of prostate cancer. The results confirm that other AL methods are 'distracted' by ambiguities and artifacts which harm the performance. FocAL effectively focuses on the most informative images, avoiding ambiguities and artifacts during acquisition. For both experiments, FocAL outperforms existing AL approaches, reaching a Cohen's kappa of 0.764 with only 0.69% of the labeled Panda data.
A General Method to Incorporate Spatial Information into Loss Functions for GAN-based Super-resolution Models
Mar 15, 2024

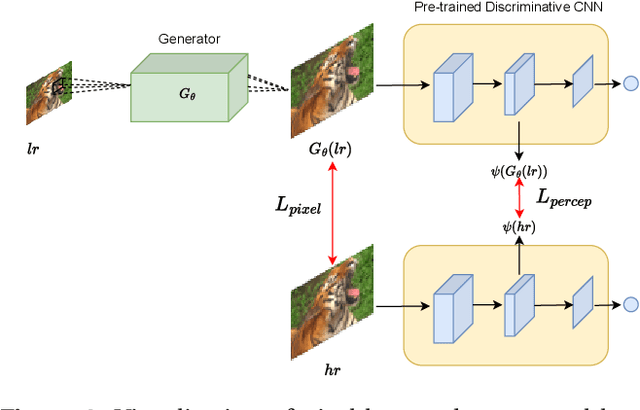
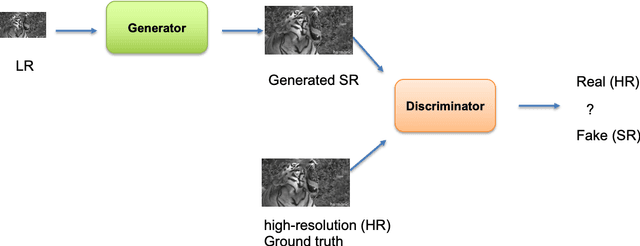
Abstract:Generative Adversarial Networks (GANs) have shown great performance on super-resolution problems since they can generate more visually realistic images and video frames. However, these models often introduce side effects into the outputs, such as unexpected artifacts and noises. To reduce these artifacts and enhance the perceptual quality of the results, in this paper, we propose a general method that can be effectively used in most GAN-based super-resolution (SR) models by introducing essential spatial information into the training process. We extract spatial information from the input data and incorporate it into the training loss, making the corresponding loss a spatially adaptive (SA) one. After that, we utilize it to guide the training process. We will show that the proposed approach is independent of the methods used to extract the spatial information and independent of the SR tasks and models. This method consistently guides the training process towards generating visually pleasing SR images and video frames, substantially mitigating artifacts and noise, ultimately leading to enhanced perceptual quality.
Introducing instance label correlation in multiple instance learning. Application to cancer detection on histopathological images
Oct 30, 2023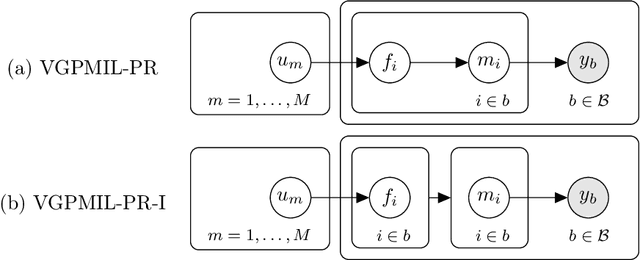
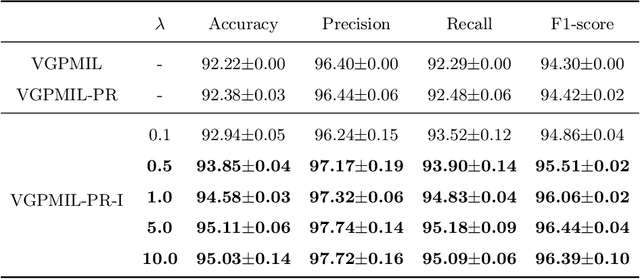
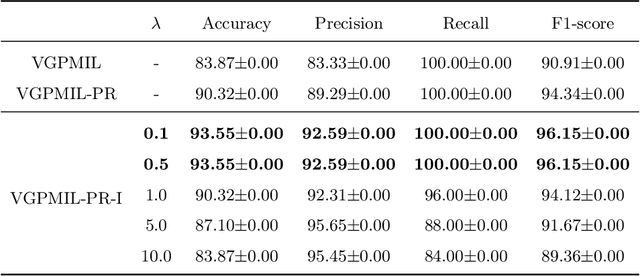
Abstract:In the last years, the weakly supervised paradigm of multiple instance learning (MIL) has become very popular in many different areas. A paradigmatic example is computational pathology, where the lack of patch-level labels for whole-slide images prevents the application of supervised models. Probabilistic MIL methods based on Gaussian Processes (GPs) have obtained promising results due to their excellent uncertainty estimation capabilities. However, these are general-purpose MIL methods that do not take into account one important fact: in (histopathological) images, the labels of neighboring patches are expected to be correlated. In this work, we extend a state-of-the-art GP-based MIL method, which is called VGPMIL-PR, to exploit such correlation. To do so, we develop a novel coupling term inspired by the statistical physics Ising model. We use variational inference to estimate all the model parameters. Interestingly, the VGPMIL-PR formulation is recovered when the weight that regulates the strength of the Ising term vanishes. The performance of the proposed method is assessed in two real-world problems of prostate cancer detection. We show that our model achieves better results than other state-of-the-art probabilistic MIL methods. We also provide different visualizations and analysis to gain insights into the influence of the novel Ising term. These insights are expected to facilitate the application of the proposed model to other research areas.
Probabilistic Modeling of Inter- and Intra-observer Variability in Medical Image Segmentation
Jul 21, 2023



Abstract:Medical image segmentation is a challenging task, particularly due to inter- and intra-observer variability, even between medical experts. In this paper, we propose a novel model, called Probabilistic Inter-Observer and iNtra-Observer variation NetwOrk (Pionono). It captures the labeling behavior of each rater with a multidimensional probability distribution and integrates this information with the feature maps of the image to produce probabilistic segmentation predictions. The model is optimized by variational inference and can be trained end-to-end. It outperforms state-of-the-art models such as STAPLE, Probabilistic U-Net, and models based on confusion matrices. Additionally, Pionono predicts multiple coherent segmentation maps that mimic the rater's expert opinion, which provides additional valuable information for the diagnostic process. Experiments on real-world cancer segmentation datasets demonstrate the high accuracy and efficiency of Pionono, making it a powerful tool for medical image analysis.
Smooth Attention for Deep Multiple Instance Learning: Application to CT Intracranial Hemorrhage Detection
Jul 18, 2023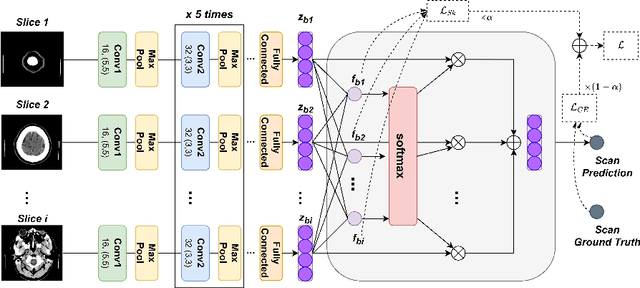

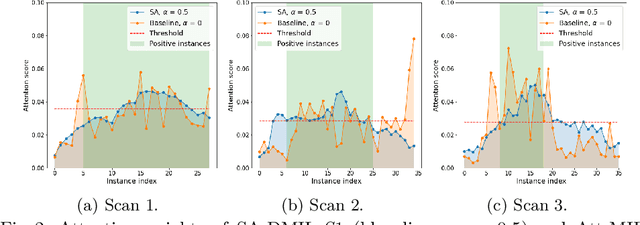
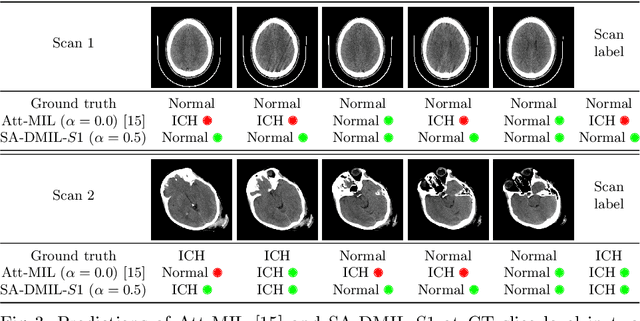
Abstract:Multiple Instance Learning (MIL) has been widely applied to medical imaging diagnosis, where bag labels are known and instance labels inside bags are unknown. Traditional MIL assumes that instances in each bag are independent samples from a given distribution. However, instances are often spatially or sequentially ordered, and one would expect similar diagnostic importance for neighboring instances. To address this, in this study, we propose a smooth attention deep MIL (SA-DMIL) model. Smoothness is achieved by the introduction of first and second order constraints on the latent function encoding the attention paid to each instance in a bag. The method is applied to the detection of intracranial hemorrhage (ICH) on head CT scans. The results show that this novel SA-DMIL: (a) achieves better performance than the non-smooth attention MIL at both scan (bag) and slice (instance) levels; (b) learns spatial dependencies between slices; and (c) outperforms current state-of-the-art MIL methods on the same ICH test set.
Probabilistic Attention based on Gaussian Processes for Deep Multiple Instance Learning
Feb 08, 2023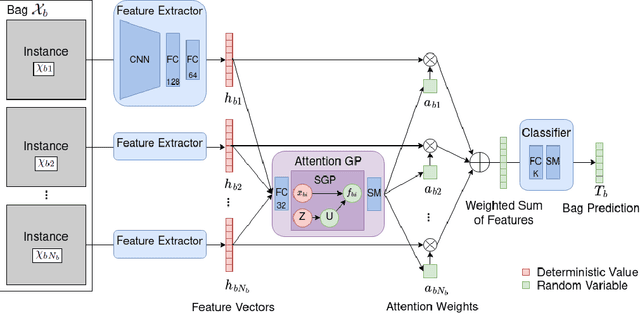
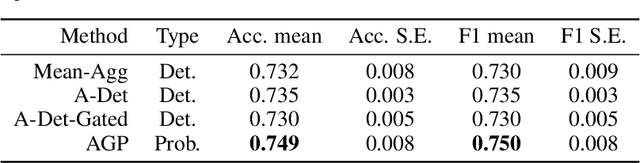
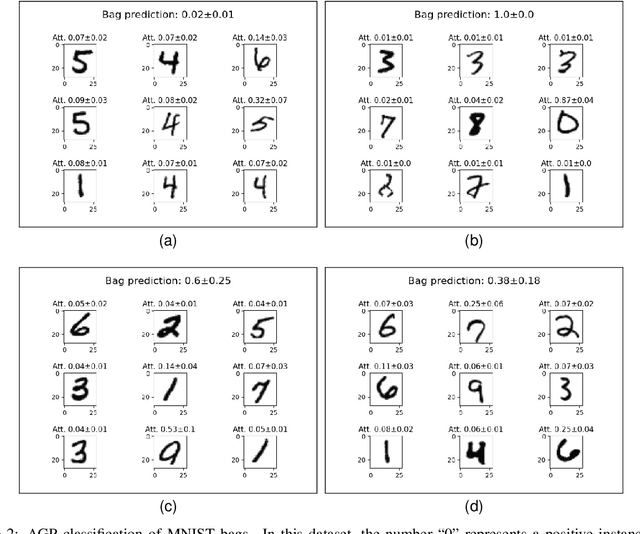
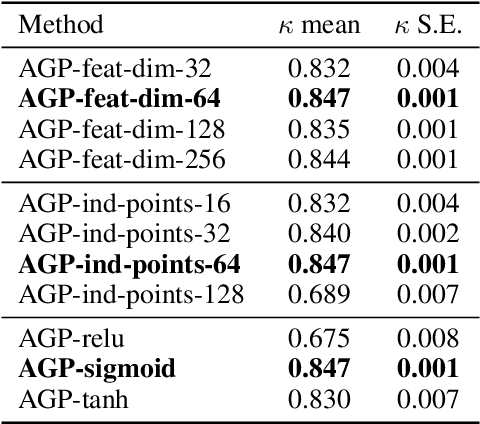
Abstract:Multiple Instance Learning (MIL) is a weakly supervised learning paradigm that is becoming increasingly popular because it requires less labeling effort than fully supervised methods. This is especially interesting for areas where the creation of large annotated datasets remains challenging, as in medicine. Although recent deep learning MIL approaches have obtained state-of-the-art results, they are fully deterministic and do not provide uncertainty estimations for the predictions. In this work, we introduce the Attention Gaussian Process (AGP) model, a novel probabilistic attention mechanism based on Gaussian Processes for deep MIL. AGP provides accurate bag-level predictions as well as instance-level explainability, and can be trained end-to-end. Moreover, its probabilistic nature guarantees robustness to overfitting on small datasets and uncertainty estimations for the predictions. The latter is especially important in medical applications, where decisions have a direct impact on the patient's health. The proposed model is validated experimentally as follows. First, its behavior is illustrated in two synthetic MIL experiments based on the well-known MNIST and CIFAR-10 datasets, respectively. Then, it is evaluated in three different real-world cancer detection experiments. AGP outperforms state-of-the-art MIL approaches, including deterministic deep learning ones. It shows a strong performance even on a small dataset with less than 100 labels and generalizes better than competing methods on an external test set. Moreover, we experimentally show that predictive uncertainty correlates with the risk of wrong predictions, and therefore it is a good indicator of reliability in practice. Our code is publicly available.
Leveraging a Probabilistic PCA Model to Understand the Multivariate Statistical Network Monitoring Framework for Network Security Anomaly Detection
Feb 02, 2023
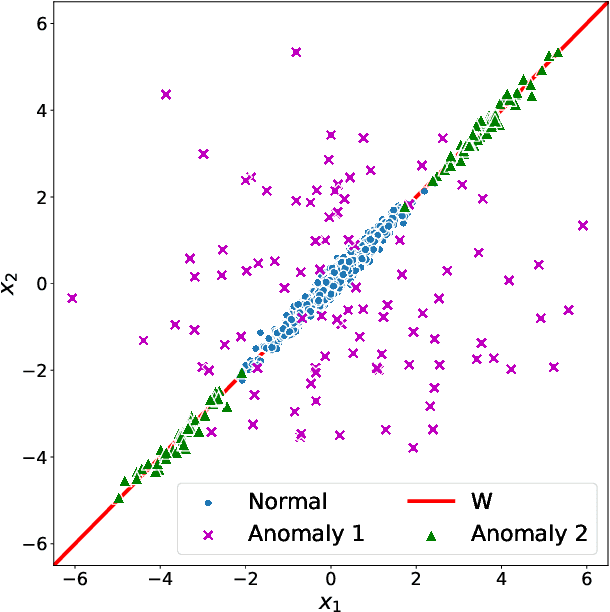
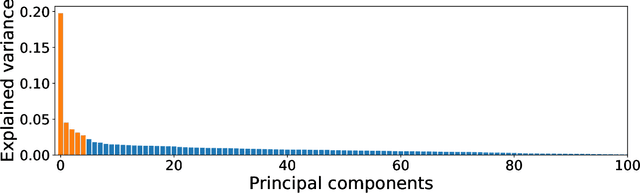
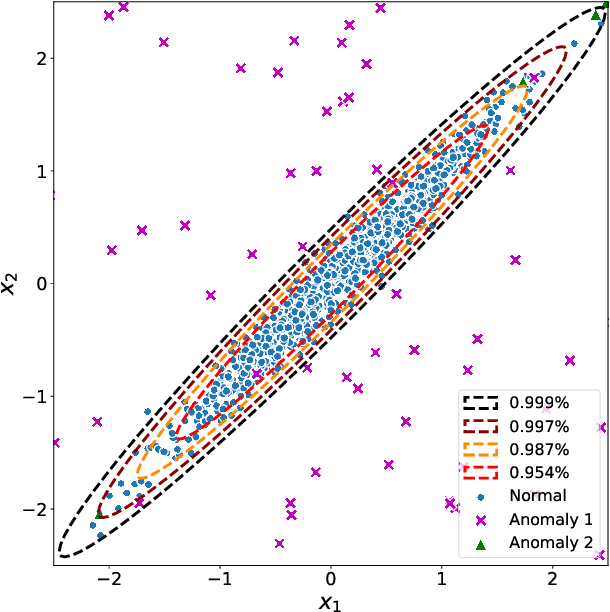
Abstract:Network anomaly detection is a very relevant research area nowadays, especially due to its multiple applications in the field of network security. The boost of new models based on variational autoencoders and generative adversarial networks has motivated a reevaluation of traditional techniques for anomaly detection. It is, however, essential to be able to understand these new models from the perspective of the experience attained from years of evaluating network security data for anomaly detection. In this paper, we revisit anomaly detection techniques based on PCA from a probabilistic generative model point of view, and contribute a mathematical model that relates them. Specifically, we start with the probabilistic PCA model and explain its connection to the Multivariate Statistical Network Monitoring (MSNM) framework. MSNM was recently successfully proposed as a means of incorporating industrial process anomaly detection experience into the field of networking. We have evaluated the mathematical model using two different datasets. The first, a synthetic dataset created to better understand the analysis proposed, and the second, UGR'16, is a specifically designed real-traffic dataset for network security anomaly detection. We have drawn conclusions that we consider to be useful when applying generative models to network security detection.
Going Deeper through the Gleason Scoring Scale: An Automatic end-to-end System for Histology Prostate Grading and Cribriform Pattern Detection
May 21, 2021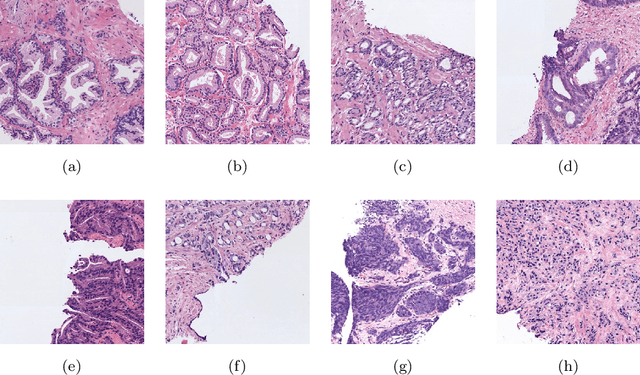



Abstract:The Gleason scoring system is the primary diagnostic and prognostic tool for prostate cancer. In recent years, with the development of digitisation devices, the use of computer vision techniques for the analysis of biopsies has increased. However, to the best of the authors' knowledge, the development of algorithms to automatically detect individual cribriform patterns belonging to Gleason grade 4 has not yet been studied in the literature. The objective of the work presented in this paper is to develop a deep-learning-based system able to support pathologists in the daily analysis of prostate biopsies. The methodological core of this work is a patch-wise predictive model based on convolutional neural networks able to determine the presence of cancerous patterns. In particular, we train from scratch a simple self-design architecture. The cribriform pattern is detected by retraining the set of filters of the last convolutional layer in the network. From the reconstructed prediction map, we compute the percentage of each Gleason grade in the tissue to feed a multi-layer perceptron which provides a biopsy-level score.mIn our SICAPv2 database, composed of 182 annotated whole slide images, we obtained a Cohen's quadratic kappa of 0.77 in the test set for the patch-level Gleason grading with the proposed architecture trained from scratch. Our results outperform previous ones reported in the literature. Furthermore, this model reaches the level of fine-tuned state-of-the-art architectures in a patient-based four groups cross validation. In the cribriform pattern detection task, we obtained an area under ROC curve of 0.82. Regarding the biopsy Gleason scoring, we achieved a quadratic Cohen's Kappa of 0.81 in the test subset. Shallow CNN architectures trained from scratch outperform current state-of-the-art methods for Gleason grades classification.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge