Lee A. D. Cooper
Department of Pathology, Northwestern University, Chicago, IL, USA
Deep Learning for Fetal Inflammatory Response Diagnosis in the Umbilical Cord
Nov 14, 2024



Abstract:Inflammation of the umbilical cord can be seen as a result of ascending intrauterine infection or other inflammatory stimuli. Acute fetal inflammatory response (FIR) is characterized by infiltration of the umbilical cord by fetal neutrophils, and can be associated with neonatal sepsis or fetal inflammatory response syndrome. Recent advances in deep learning in digital pathology have demonstrated favorable performance across a wide range of clinical tasks, such as diagnosis and prognosis. In this study we classified FIR from whole slide images (WSI). We digitized 4100 histological slides of umbilical cord stained with hematoxylin and eosin(H&E) and extracted placental diagnoses from the electronic health record. We build models using attention-based whole slide learning models. We compared strategies between features extracted by a model (ConvNeXtXLarge) pretrained on non-medical images (ImageNet), and one pretrained using histopathology images (UNI). We trained multiple iterations of each model and combined them into an ensemble. The predictions from the ensemble of models trained using UNI achieved an overall balanced accuracy of 0.836 on the test dataset. In comparison, the ensembled predictions using ConvNeXtXLarge had a lower balanced accuracy of 0.7209. Heatmaps generated from top accuracy model appropriately highlighted arteritis in cases of FIR 2. In FIR 1, the highest performing model assigned high attention to areas of activated-appearing stroma in Wharton's Jelly. However, other high-performing models assigned attention to umbilical vessels. We developed models for diagnosis of FIR from placental histology images, helping reduce interobserver variability among pathologists. Future work may examine the utility of these models for identifying infants at risk of systemic inflammatory response or early onset neonatal sepsis.
Machine learning identification of maternal inflammatory response and histologic choroamnionitis from placental membrane whole slide images
Nov 04, 2024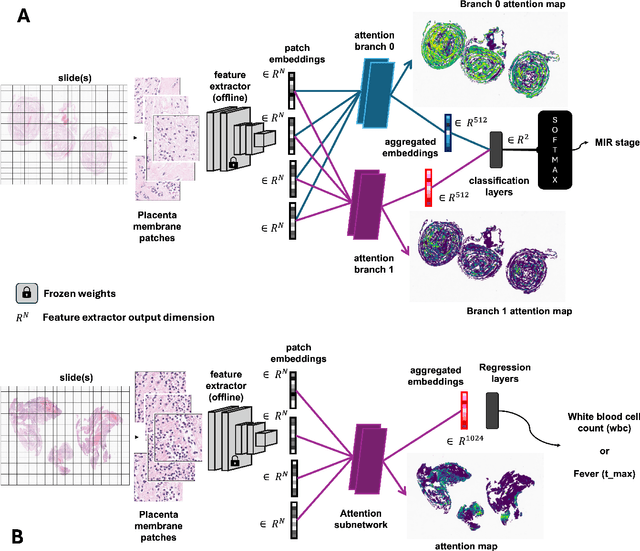

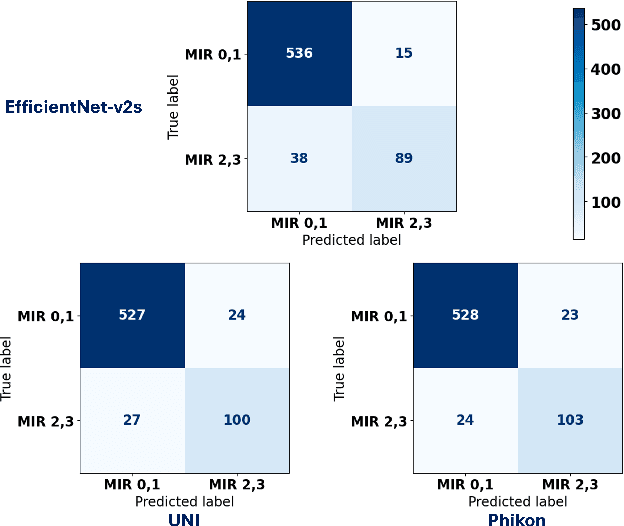
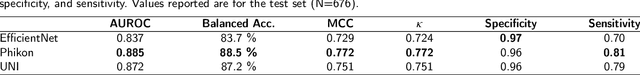
Abstract:The placenta forms a critical barrier to infection through pregnancy, labor and, delivery. Inflammatory processes in the placenta have short-term, and long-term consequences for offspring health. Digital pathology and machine learning can play an important role in understanding placental inflammation, and there have been very few investigations into methods for predicting and understanding Maternal Inflammatory Response (MIR). This work intends to investigate the potential of using machine learning to understand MIR based on whole slide images (WSI), and establish early benchmarks. To that end, we use Multiple Instance Learning framework with 3 feature extractors: ImageNet-based EfficientNet-v2s, and 2 histopathology foundation models, UNI and Phikon to investigate predictability of MIR stage from histopathology WSIs. We also interpret predictions from these models using the learned attention maps from these models. We also use the MIL framework for predicting white blood cells count (WBC) and maximum fever temperature ($T_{max}$). Attention-based MIL models are able to classify MIR with a balanced accuracy of up to 88.5% with a Cohen's Kappa ($\kappa$) of up to 0.772. Furthermore, we found that the pathology foundation models (UNI and Phikon) are both able to achieve higher performance with balanced accuracy and $\kappa$, compared to ImageNet-based feature extractor (EfficientNet-v2s). For WBC and $T_{max}$ prediction, we found mild correlation between actual values and those predicted from histopathology WSIs. We used MIL framework for predicting MIR stage from WSIs, and compared effectiveness of foundation models as feature extractors, with that of an ImageNet-based model. We further investigated model failure cases and found them to be either edge cases prone to interobserver variability, examples of pathologist's overreach, or mislabeled due to processing errors.
BraTS-Path Challenge: Assessing Heterogeneous Histopathologic Brain Tumor Sub-regions
May 17, 2024Abstract:Glioblastoma is the most common primary adult brain tumor, with a grim prognosis - median survival of 12-18 months following treatment, and 4 months otherwise. Glioblastoma is widely infiltrative in the cerebral hemispheres and well-defined by heterogeneous molecular and micro-environmental histopathologic profiles, which pose a major obstacle in treatment. Correctly diagnosing these tumors and assessing their heterogeneity is crucial for choosing the precise treatment and potentially enhancing patient survival rates. In the gold-standard histopathology-based approach to tumor diagnosis, detecting various morpho-pathological features of distinct histology throughout digitized tissue sections is crucial. Such "features" include the presence of cellular tumor, geographic necrosis, pseudopalisading necrosis, areas abundant in microvascular proliferation, infiltration into the cortex, wide extension in subcortical white matter, leptomeningeal infiltration, regions dense with macrophages, and the presence of perivascular or scattered lymphocytes. With these features in mind and building upon the main aim of the BraTS Cluster of Challenges https://www.synapse.org/brats2024, the goal of the BraTS-Path challenge is to provide a systematically prepared comprehensive dataset and a benchmarking environment to develop and fairly compare deep-learning models capable of identifying tumor sub-regions of distinct histologic profile. These models aim to further our understanding of the disease and assist in the diagnosis and grading of conditions in a consistent manner.
Focused Active Learning for Histopathological Image Classification
Apr 06, 2024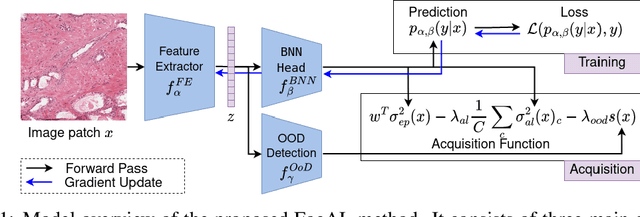
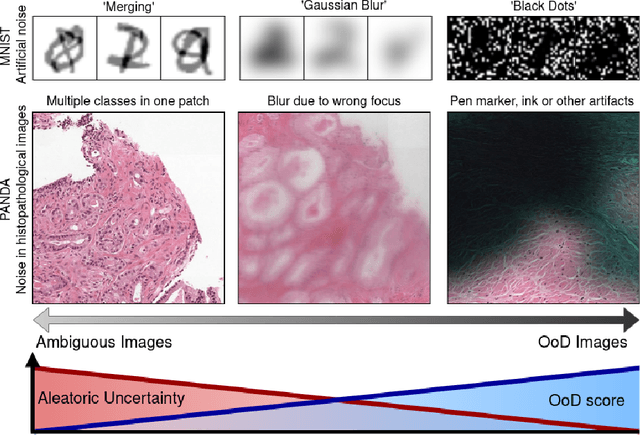
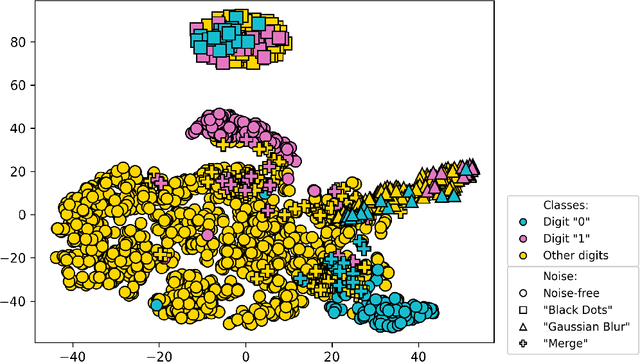
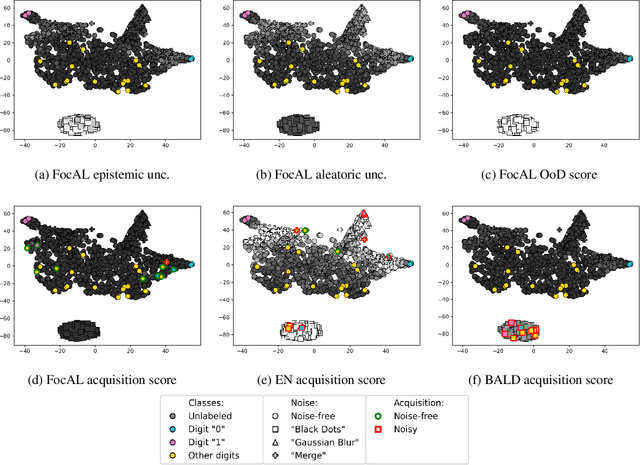
Abstract:Active Learning (AL) has the potential to solve a major problem of digital pathology: the efficient acquisition of labeled data for machine learning algorithms. However, existing AL methods often struggle in realistic settings with artifacts, ambiguities, and class imbalances, as commonly seen in the medical field. The lack of precise uncertainty estimations leads to the acquisition of images with a low informative value. To address these challenges, we propose Focused Active Learning (FocAL), which combines a Bayesian Neural Network with Out-of-Distribution detection to estimate different uncertainties for the acquisition function. Specifically, the weighted epistemic uncertainty accounts for the class imbalance, aleatoric uncertainty for ambiguous images, and an OoD score for artifacts. We perform extensive experiments to validate our method on MNIST and the real-world Panda dataset for the classification of prostate cancer. The results confirm that other AL methods are 'distracted' by ambiguities and artifacts which harm the performance. FocAL effectively focuses on the most informative images, avoiding ambiguities and artifacts during acquisition. For both experiments, FocAL outperforms existing AL approaches, reaching a Cohen's kappa of 0.764 with only 0.69% of the labeled Panda data.
MONAI: An open-source framework for deep learning in healthcare
Nov 04, 2022



Abstract:Artificial Intelligence (AI) is having a tremendous impact across most areas of science. Applications of AI in healthcare have the potential to improve our ability to detect, diagnose, prognose, and intervene on human disease. For AI models to be used clinically, they need to be made safe, reproducible and robust, and the underlying software framework must be aware of the particularities (e.g. geometry, physiology, physics) of medical data being processed. This work introduces MONAI, a freely available, community-supported, and consortium-led PyTorch-based framework for deep learning in healthcare. MONAI extends PyTorch to support medical data, with a particular focus on imaging, and provide purpose-specific AI model architectures, transformations and utilities that streamline the development and deployment of medical AI models. MONAI follows best practices for software-development, providing an easy-to-use, robust, well-documented, and well-tested software framework. MONAI preserves the simple, additive, and compositional approach of its underlying PyTorch libraries. MONAI is being used by and receiving contributions from research, clinical and industrial teams from around the world, who are pursuing applications spanning nearly every aspect of healthcare.
A Histopathology Study Comparing Contrastive Semi-Supervised and Fully Supervised Learning
Nov 10, 2021
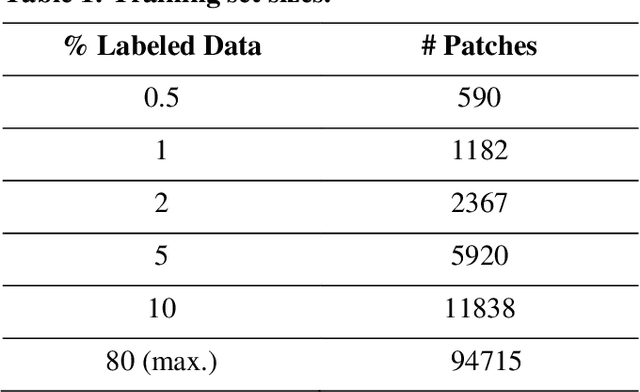
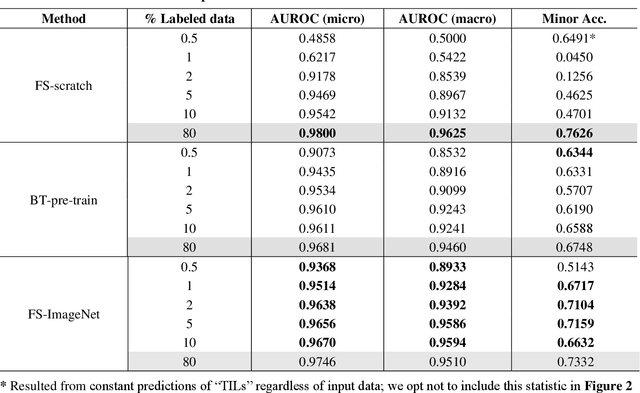
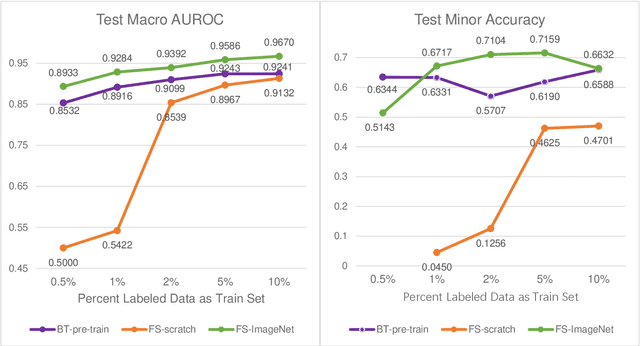
Abstract:Data labeling is often the most challenging task when developing computational pathology models. Pathologist participation is necessary to generate accurate labels, and the limitations on pathologist time and demand for large, labeled datasets has led to research in areas including weakly supervised learning using patient-level labels, machine assisted annotation and active learning. In this paper we explore self-supervised learning to reduce labeling burdens in computational pathology. We explore this in the context of classification of breast cancer tissue using the Barlow Twins approach, and we compare self-supervision with alternatives like pre-trained networks in low-data scenarios. For the task explored in this paper, we find that ImageNet pre-trained networks largely outperform the self-supervised representations obtained using Barlow Twins.
NuCLS: A scalable crowdsourcing, deep learning approach and dataset for nucleus classification, localization and segmentation
Feb 18, 2021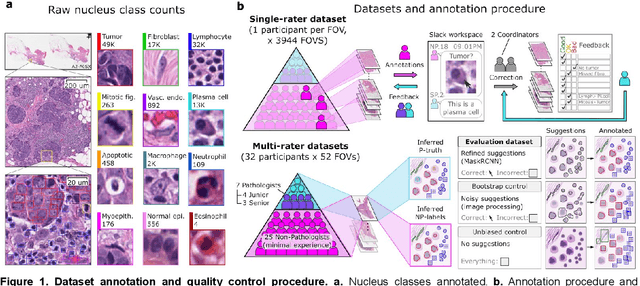
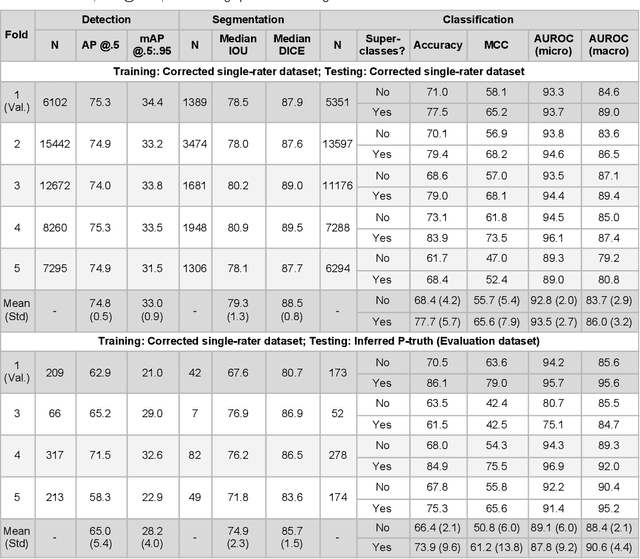
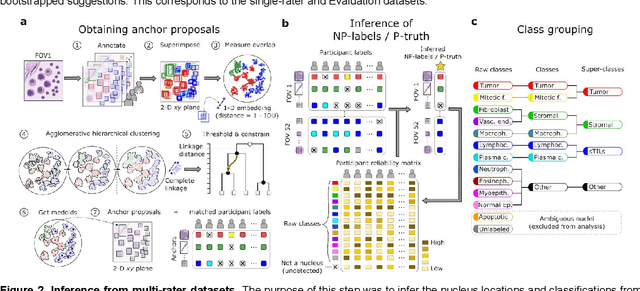
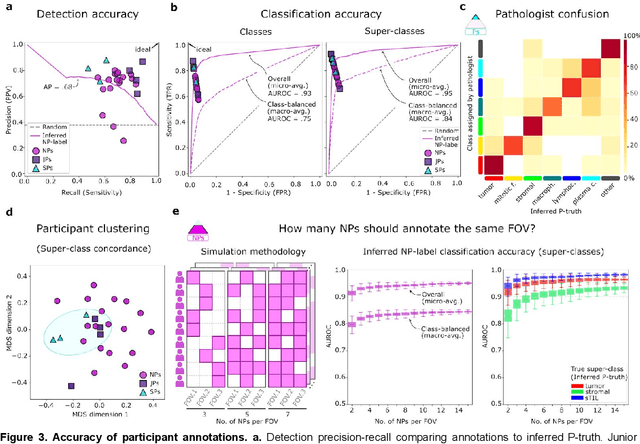
Abstract:High-resolution mapping of cells and tissue structures provides a foundation for developing interpretable machine-learning models for computational pathology. Deep learning algorithms can provide accurate mappings given large numbers of labeled instances for training and validation. Generating adequate volume of quality labels has emerged as a critical barrier in computational pathology given the time and effort required from pathologists. In this paper we describe an approach for engaging crowds of medical students and pathologists that was used to produce a dataset of over 220,000 annotations of cell nuclei in breast cancers. We show how suggested annotations generated by a weak algorithm can improve the accuracy of annotations generated by non-experts and can yield useful data for training segmentation algorithms without laborious manual tracing. We systematically examine interrater agreement and describe modifications to the MaskRCNN model to improve cell mapping. We also describe a technique we call Decision Tree Approximation of Learned Embeddings (DTALE) that leverages nucleus segmentations and morphologic features to improve the transparency of nucleus classification models. The annotation data produced in this study are freely available for algorithm development and benchmarking at: https://sites.google.com/view/nucls.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge