Machine learning identification of maternal inflammatory response and histologic choroamnionitis from placental membrane whole slide images
Paper and Code
Nov 04, 2024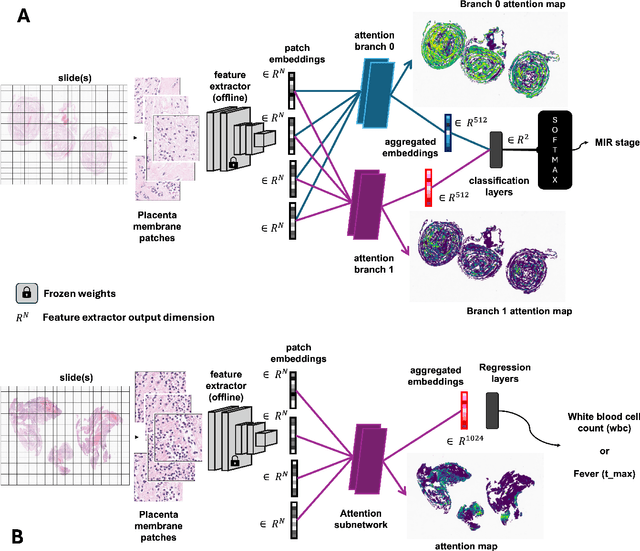

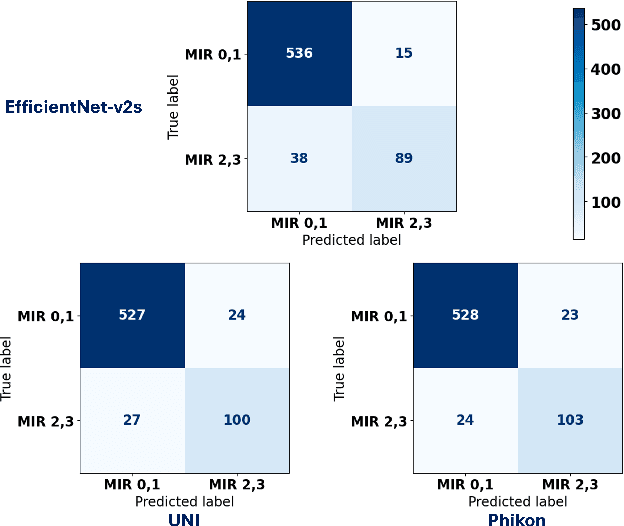
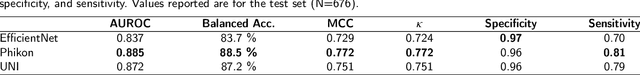
The placenta forms a critical barrier to infection through pregnancy, labor and, delivery. Inflammatory processes in the placenta have short-term, and long-term consequences for offspring health. Digital pathology and machine learning can play an important role in understanding placental inflammation, and there have been very few investigations into methods for predicting and understanding Maternal Inflammatory Response (MIR). This work intends to investigate the potential of using machine learning to understand MIR based on whole slide images (WSI), and establish early benchmarks. To that end, we use Multiple Instance Learning framework with 3 feature extractors: ImageNet-based EfficientNet-v2s, and 2 histopathology foundation models, UNI and Phikon to investigate predictability of MIR stage from histopathology WSIs. We also interpret predictions from these models using the learned attention maps from these models. We also use the MIL framework for predicting white blood cells count (WBC) and maximum fever temperature ($T_{max}$). Attention-based MIL models are able to classify MIR with a balanced accuracy of up to 88.5% with a Cohen's Kappa ($\kappa$) of up to 0.772. Furthermore, we found that the pathology foundation models (UNI and Phikon) are both able to achieve higher performance with balanced accuracy and $\kappa$, compared to ImageNet-based feature extractor (EfficientNet-v2s). For WBC and $T_{max}$ prediction, we found mild correlation between actual values and those predicted from histopathology WSIs. We used MIL framework for predicting MIR stage from WSIs, and compared effectiveness of foundation models as feature extractors, with that of an ImageNet-based model. We further investigated model failure cases and found them to be either edge cases prone to interobserver variability, examples of pathologist's overreach, or mislabeled due to processing errors.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge