Ramin Nateghi
Deep Learning for Fetal Inflammatory Response Diagnosis in the Umbilical Cord
Nov 14, 2024



Abstract:Inflammation of the umbilical cord can be seen as a result of ascending intrauterine infection or other inflammatory stimuli. Acute fetal inflammatory response (FIR) is characterized by infiltration of the umbilical cord by fetal neutrophils, and can be associated with neonatal sepsis or fetal inflammatory response syndrome. Recent advances in deep learning in digital pathology have demonstrated favorable performance across a wide range of clinical tasks, such as diagnosis and prognosis. In this study we classified FIR from whole slide images (WSI). We digitized 4100 histological slides of umbilical cord stained with hematoxylin and eosin(H&E) and extracted placental diagnoses from the electronic health record. We build models using attention-based whole slide learning models. We compared strategies between features extracted by a model (ConvNeXtXLarge) pretrained on non-medical images (ImageNet), and one pretrained using histopathology images (UNI). We trained multiple iterations of each model and combined them into an ensemble. The predictions from the ensemble of models trained using UNI achieved an overall balanced accuracy of 0.836 on the test dataset. In comparison, the ensembled predictions using ConvNeXtXLarge had a lower balanced accuracy of 0.7209. Heatmaps generated from top accuracy model appropriately highlighted arteritis in cases of FIR 2. In FIR 1, the highest performing model assigned high attention to areas of activated-appearing stroma in Wharton's Jelly. However, other high-performing models assigned attention to umbilical vessels. We developed models for diagnosis of FIR from placental histology images, helping reduce interobserver variability among pathologists. Future work may examine the utility of these models for identifying infants at risk of systemic inflammatory response or early onset neonatal sepsis.
Machine learning identification of maternal inflammatory response and histologic choroamnionitis from placental membrane whole slide images
Nov 04, 2024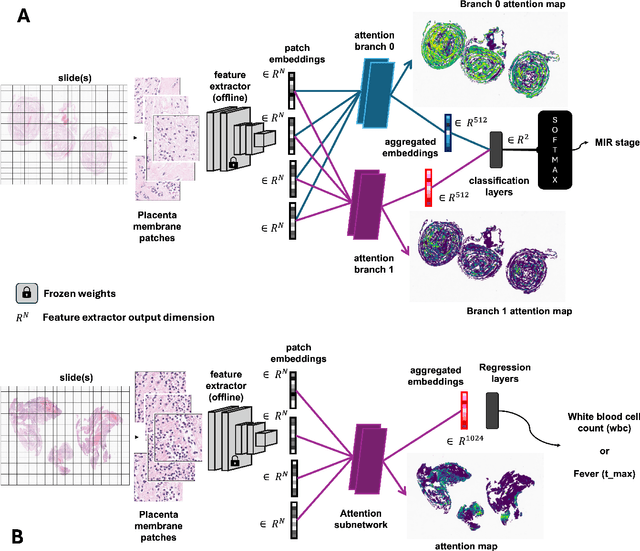

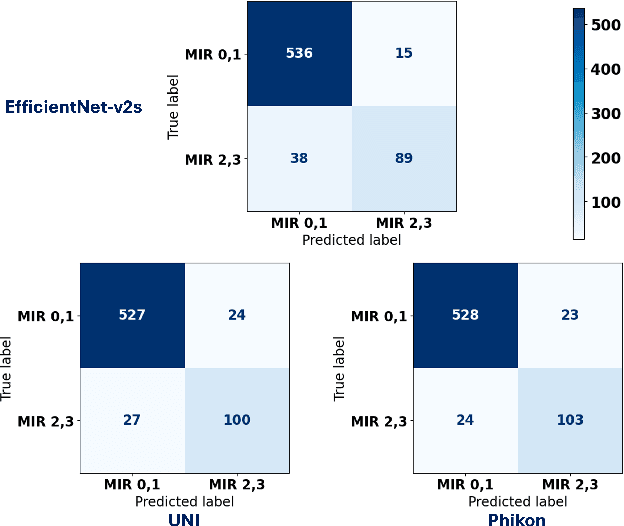
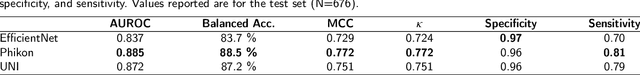
Abstract:The placenta forms a critical barrier to infection through pregnancy, labor and, delivery. Inflammatory processes in the placenta have short-term, and long-term consequences for offspring health. Digital pathology and machine learning can play an important role in understanding placental inflammation, and there have been very few investigations into methods for predicting and understanding Maternal Inflammatory Response (MIR). This work intends to investigate the potential of using machine learning to understand MIR based on whole slide images (WSI), and establish early benchmarks. To that end, we use Multiple Instance Learning framework with 3 feature extractors: ImageNet-based EfficientNet-v2s, and 2 histopathology foundation models, UNI and Phikon to investigate predictability of MIR stage from histopathology WSIs. We also interpret predictions from these models using the learned attention maps from these models. We also use the MIL framework for predicting white blood cells count (WBC) and maximum fever temperature ($T_{max}$). Attention-based MIL models are able to classify MIR with a balanced accuracy of up to 88.5% with a Cohen's Kappa ($\kappa$) of up to 0.772. Furthermore, we found that the pathology foundation models (UNI and Phikon) are both able to achieve higher performance with balanced accuracy and $\kappa$, compared to ImageNet-based feature extractor (EfficientNet-v2s). For WBC and $T_{max}$ prediction, we found mild correlation between actual values and those predicted from histopathology WSIs. We used MIL framework for predicting MIR stage from WSIs, and compared effectiveness of foundation models as feature extractors, with that of an ImageNet-based model. We further investigated model failure cases and found them to be either edge cases prone to interobserver variability, examples of pathologist's overreach, or mislabeled due to processing errors.
Novel Clinical-Grade Prostate Cancer Detection and Grading Model: Development and Prospective Validation Using Real World Data, with Performance Assessment on IHC Requested Cases
Oct 31, 2024



Abstract:Artificial intelligence may assist healthcare systems in meeting increasing demand for pathology services while maintaining diagnostic quality and reducing turnaround time and costs. We aimed to investigate the performance of an institutionally developed system for prostate cancer detection, grading, and workflow optimization and to contrast this with commercial alternatives. From August 2021 to March 2023, we scanned 21,396 slides from 1,147 patients with positive biopsies. We developed models for cancer detection, grading, and screening of equivocal cases for IHC ordering. We compared a task-specific model trained using the PANDA dataset of prostate cancer biopsies with one built using features extracted by the general-purpose histology foundation model, UNI and compare their performance in an unfiltered prospectively collected dataset that reflects our patient population (1737 slides,95 patients). We evaluated the contributions of a bespoke model designed to improve sensitivity in detecting small cancer foci and scoring of broader patterns observed at lower resolution. We found high concordance between the developed systems and pathologist reference in detection (AUC 98.5, sensitivity 95.0, and specificity 97.8), ISUP grading (quadratic Cohen's kappa 0.869), grade group 3 or higher (AUC 97.5, sensitivity 94.9, specificity 96.6) and comparable to published data from commercial systems. Screening could reduce IHC ordering for equivocal cases by 44.5% with an overall error rate of 1.8% (1.4% false positive, 0.4% false negative rates). Institutions like academic medical centers that have high scanning volumes and report abstraction capabilities can develop accurate computational pathology models for internal use. These models have the potential to aid in quality control role and to improve workflow in the pathology lab to help meet future challenges in prostate cancer diagnosis.
Biomedical image analysis competitions: The state of current participation practice
Dec 16, 2022Abstract:The number of international benchmarking competitions is steadily increasing in various fields of machine learning (ML) research and practice. So far, however, little is known about the common practice as well as bottlenecks faced by the community in tackling the research questions posed. To shed light on the status quo of algorithm development in the specific field of biomedical imaging analysis, we designed an international survey that was issued to all participants of challenges conducted in conjunction with the IEEE ISBI 2021 and MICCAI 2021 conferences (80 competitions in total). The survey covered participants' expertise and working environments, their chosen strategies, as well as algorithm characteristics. A median of 72% challenge participants took part in the survey. According to our results, knowledge exchange was the primary incentive (70%) for participation, while the reception of prize money played only a minor role (16%). While a median of 80 working hours was spent on method development, a large portion of participants stated that they did not have enough time for method development (32%). 25% perceived the infrastructure to be a bottleneck. Overall, 94% of all solutions were deep learning-based. Of these, 84% were based on standard architectures. 43% of the respondents reported that the data samples (e.g., images) were too large to be processed at once. This was most commonly addressed by patch-based training (69%), downsampling (37%), and solving 3D analysis tasks as a series of 2D tasks. K-fold cross-validation on the training set was performed by only 37% of the participants and only 50% of the participants performed ensembling based on multiple identical models (61%) or heterogeneous models (39%). 48% of the respondents applied postprocessing steps.
Mitosis domain generalization in histopathology images -- The MIDOG challenge
Apr 06, 2022
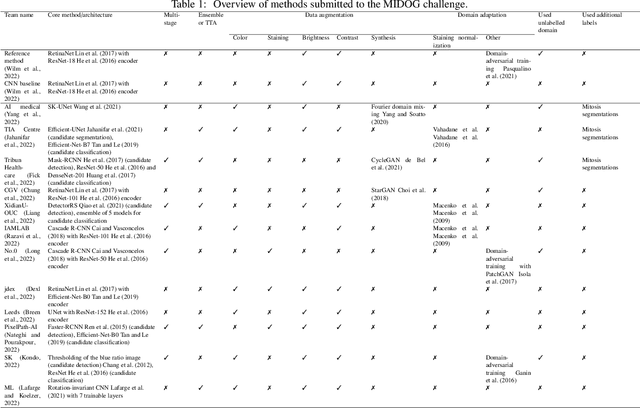
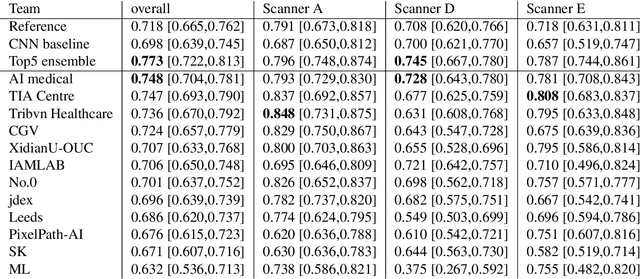
Abstract:The density of mitotic figures within tumor tissue is known to be highly correlated with tumor proliferation and thus is an important marker in tumor grading. Recognition of mitotic figures by pathologists is known to be subject to a strong inter-rater bias, which limits the prognostic value. State-of-the-art deep learning methods can support the expert in this assessment but are known to strongly deteriorate when applied in a different clinical environment than was used for training. One decisive component in the underlying domain shift has been identified as the variability caused by using different whole slide scanners. The goal of the MICCAI MIDOG 2021 challenge has been to propose and evaluate methods that counter this domain shift and derive scanner-agnostic mitosis detection algorithms. The challenge used a training set of 200 cases, split across four scanning systems. As a test set, an additional 100 cases split across four scanning systems, including two previously unseen scanners, were given. The best approaches performed on an expert level, with the winning algorithm yielding an F_1 score of 0.748 (CI95: 0.704-0.781). In this paper, we evaluate and compare the approaches that were submitted to the challenge and identify methodological factors contributing to better performance.
Perineural Invasion Detection in Multiple Organ Cancer Based on Deep Convolutional Neural Network
Oct 23, 2021



Abstract:Perineural invasion (PNI) by malignant tumor cells has been reported as an independent indicator of poor prognosis in various cancers. Assessment of PNI in small nerves on glass slides is a labor-intensive task. In this study, we propose an algorithm to detect the perineural invasions in colon, prostate, and pancreas cancers based on a convolutional neural network (CNN).
Two-step Domain Adaptation for Mitosis Cell Detection in Histopathology Images
Aug 31, 2021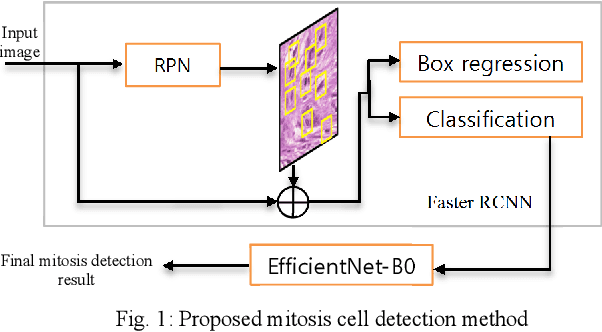
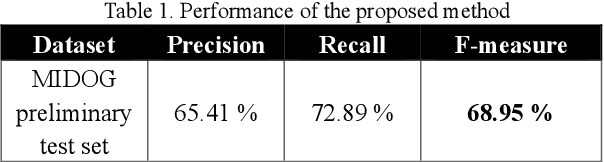
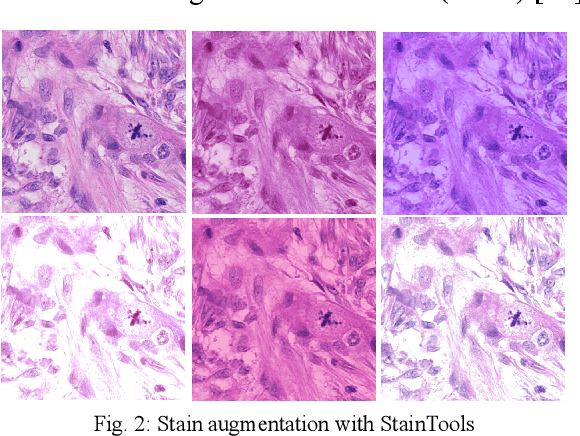
Abstract:We propose a two-step domain shift-invariant mitosis cell detection method based on Faster RCNN and a convolutional neural network (CNN). We generate various domain-shifted versions of existing histopathology images using a stain augmentation technique, enabling our method to effectively learn various stain domains and achieve better generalization. The performance of our method is evaluated on the preliminary test data set of the MIDOG-2021 challenge. The experimental results demonstrate that the proposed mitosis detection method can achieve promising performance for domain-shifted histopathology images.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge