Lawrence Chillrud
Department of Environmental Health Sciences, Columbia University Mailman School of Public Health
Deep Learning for Fetal Inflammatory Response Diagnosis in the Umbilical Cord
Nov 14, 2024



Abstract:Inflammation of the umbilical cord can be seen as a result of ascending intrauterine infection or other inflammatory stimuli. Acute fetal inflammatory response (FIR) is characterized by infiltration of the umbilical cord by fetal neutrophils, and can be associated with neonatal sepsis or fetal inflammatory response syndrome. Recent advances in deep learning in digital pathology have demonstrated favorable performance across a wide range of clinical tasks, such as diagnosis and prognosis. In this study we classified FIR from whole slide images (WSI). We digitized 4100 histological slides of umbilical cord stained with hematoxylin and eosin(H&E) and extracted placental diagnoses from the electronic health record. We build models using attention-based whole slide learning models. We compared strategies between features extracted by a model (ConvNeXtXLarge) pretrained on non-medical images (ImageNet), and one pretrained using histopathology images (UNI). We trained multiple iterations of each model and combined them into an ensemble. The predictions from the ensemble of models trained using UNI achieved an overall balanced accuracy of 0.836 on the test dataset. In comparison, the ensembled predictions using ConvNeXtXLarge had a lower balanced accuracy of 0.7209. Heatmaps generated from top accuracy model appropriately highlighted arteritis in cases of FIR 2. In FIR 1, the highest performing model assigned high attention to areas of activated-appearing stroma in Wharton's Jelly. However, other high-performing models assigned attention to umbilical vessels. We developed models for diagnosis of FIR from placental histology images, helping reduce interobserver variability among pathologists. Future work may examine the utility of these models for identifying infants at risk of systemic inflammatory response or early onset neonatal sepsis.
Check-COVID: Fact-Checking COVID-19 News Claims with Scientific Evidence
May 29, 2023



Abstract:We present a new fact-checking benchmark, Check-COVID, that requires systems to verify claims about COVID-19 from news using evidence from scientific articles. This approach to fact-checking is particularly challenging as it requires checking internet text written in everyday language against evidence from journal articles written in formal academic language. Check-COVID contains 1, 504 expert-annotated news claims about the coronavirus paired with sentence-level evidence from scientific journal articles and veracity labels. It includes both extracted (journalist-written) and composed (annotator-written) claims. Experiments using both a fact-checking specific system and GPT-3.5, which respectively achieve F1 scores of 76.99 and 69.90 on this task, reveal the difficulty of automatically fact-checking both claim types and the importance of in-domain data for good performance. Our data and models are released publicly at https://github.com/posuer/Check-COVID.
Principal Component Pursuit for Pattern Identification in Environmental Mixtures
Oct 29, 2021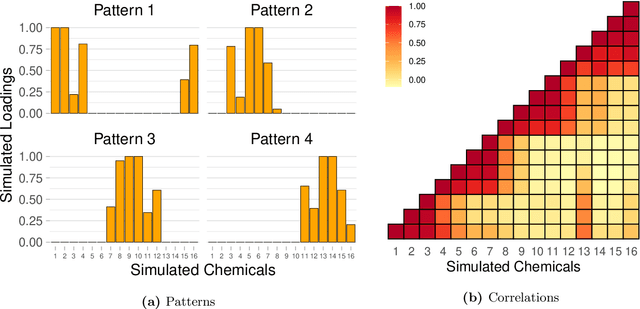
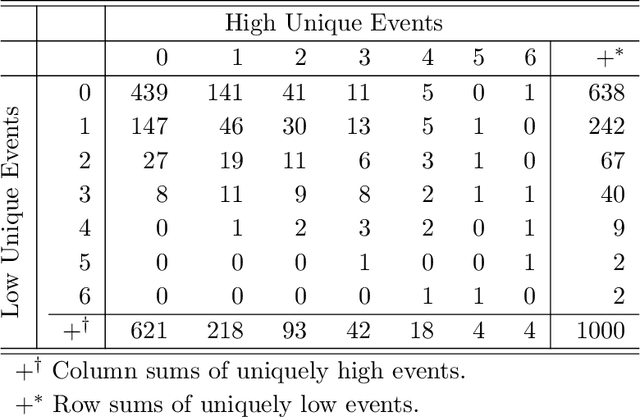
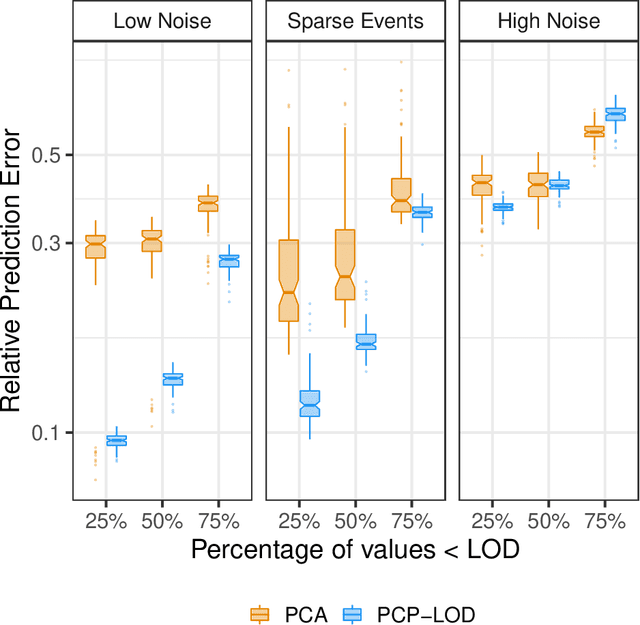
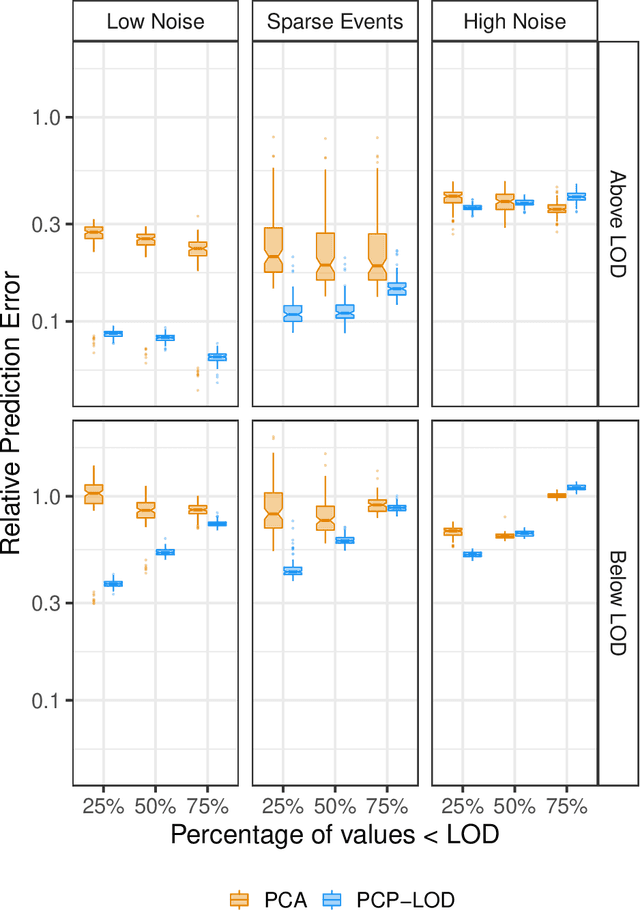
Abstract:Environmental health researchers often aim to identify sources/behaviors that give rise to potentially harmful exposures. We adapted principal component pursuit (PCP)-a robust technique for dimensionality reduction in computer vision and signal processing-to identify patterns in environmental mixtures. PCP decomposes the exposure mixture into a low-rank matrix containing consistent exposure patterns across pollutants and a sparse matrix isolating unique exposure events. We adapted PCP to accommodate non-negative and missing data, and values below a given limit of detection (LOD). We simulated data to represent environmental mixtures of two sizes with increasing proportions <LOD and three noise structures. We compared PCP-LOD to principal component analysis (PCA) to evaluate performance. We next applied PCP-LOD to a mixture of 21 persistent organic pollutants (POPs) measured in 1,000 U.S. adults from the 2001-2002 National Health and Nutrition Examination Survey. We applied singular value decomposition to the estimated low-rank matrix to characterize the patterns. PCP-LOD recovered the true number of patterns through cross-validation for all simulations; based on an a priori specified criterion, PCA recovered the true number of patterns in 32% of simulations. PCP-LOD achieved lower relative predictive error than PCA for all simulated datasets with up to 50% of the data <LOD. When 75% of values were <LOD, PCP-LOD outperformed PCA only when noise was low. In the POP mixture, PCP-LOD identified a rank-three underlying structure and separated 6% of values as unique events. One pattern represented comprehensive exposure to all POPs. The other patterns grouped chemicals based on known structure and toxicity. PCP-LOD serves as a useful tool to express multi-dimensional exposures as consistent patterns that, if found to be related to adverse health, are amenable to targeted interventions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge