Julie B. Herbstman
Department of Environmental Health Sciences, Columbia University Mailman School of Public Health
Principal Component Pursuit for Pattern Identification in Environmental Mixtures
Oct 29, 2021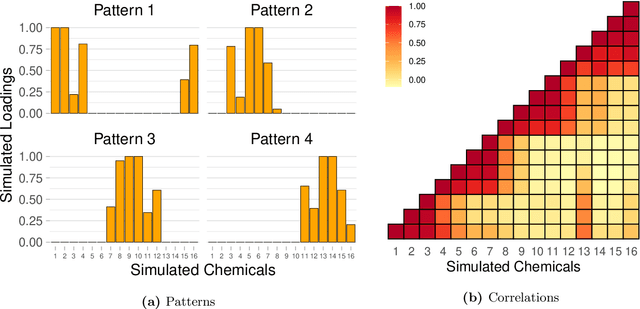
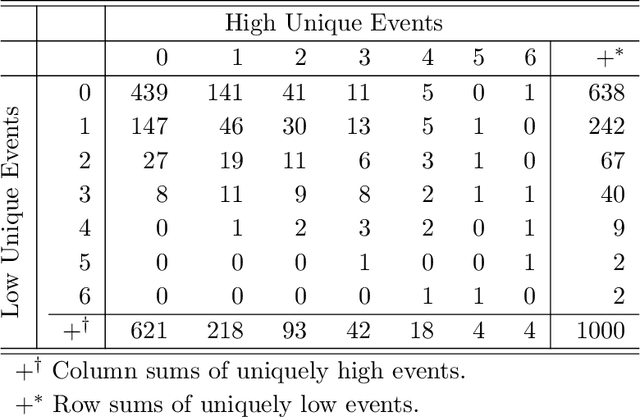
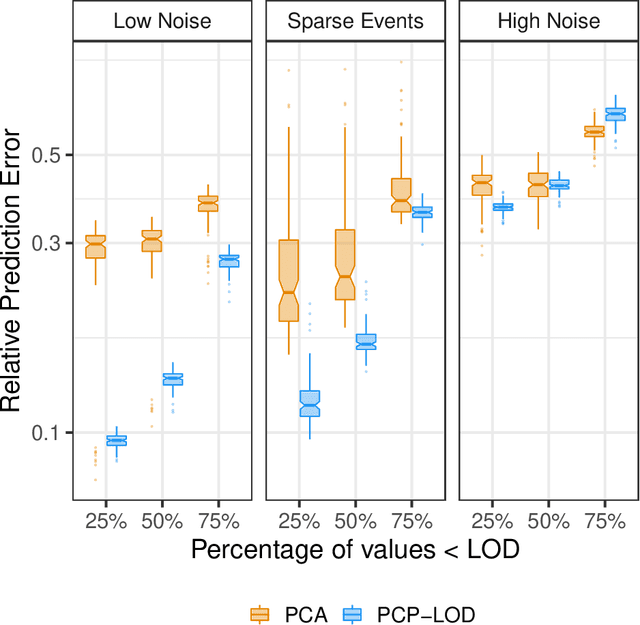
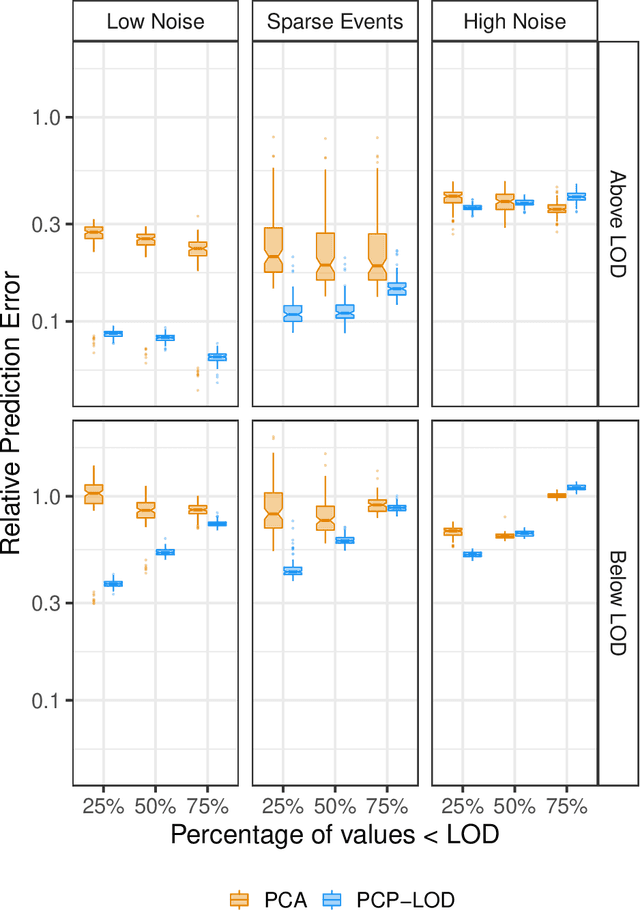
Abstract:Environmental health researchers often aim to identify sources/behaviors that give rise to potentially harmful exposures. We adapted principal component pursuit (PCP)-a robust technique for dimensionality reduction in computer vision and signal processing-to identify patterns in environmental mixtures. PCP decomposes the exposure mixture into a low-rank matrix containing consistent exposure patterns across pollutants and a sparse matrix isolating unique exposure events. We adapted PCP to accommodate non-negative and missing data, and values below a given limit of detection (LOD). We simulated data to represent environmental mixtures of two sizes with increasing proportions <LOD and three noise structures. We compared PCP-LOD to principal component analysis (PCA) to evaluate performance. We next applied PCP-LOD to a mixture of 21 persistent organic pollutants (POPs) measured in 1,000 U.S. adults from the 2001-2002 National Health and Nutrition Examination Survey. We applied singular value decomposition to the estimated low-rank matrix to characterize the patterns. PCP-LOD recovered the true number of patterns through cross-validation for all simulations; based on an a priori specified criterion, PCA recovered the true number of patterns in 32% of simulations. PCP-LOD achieved lower relative predictive error than PCA for all simulated datasets with up to 50% of the data <LOD. When 75% of values were <LOD, PCP-LOD outperformed PCA only when noise was low. In the POP mixture, PCP-LOD identified a rank-three underlying structure and separated 6% of values as unique events. One pattern represented comprehensive exposure to all POPs. The other patterns grouped chemicals based on known structure and toxicity. PCP-LOD serves as a useful tool to express multi-dimensional exposures as consistent patterns that, if found to be related to adverse health, are amenable to targeted interventions.
Bayesian non-parametric non-negative matrix factorization for pattern identification in environmental mixtures
Sep 24, 2021



Abstract:Environmental health researchers may aim to identify exposure patterns that represent sources, product use, or behaviors that give rise to mixtures of potentially harmful environmental chemical exposures. We present Bayesian non-parametric non-negative matrix factorization (BN^2MF) as a novel method to identify patterns of chemical exposures when the number of patterns is not known a priori. We placed non-negative continuous priors on pattern loadings and individual scores to enhance interpretability and used a clever non-parametric sparse prior to estimate the pattern number. We further derived variational confidence intervals around estimates; this is a critical development because it quantifies the model's confidence in estimated patterns. These unique features contrast with existing pattern recognition methods employed in this field which are limited by user-specified pattern number, lack of interpretability of patterns in terms of human understanding, and lack of uncertainty quantification.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge