A Histopathology Study Comparing Contrastive Semi-Supervised and Fully Supervised Learning
Paper and Code
Nov 10, 2021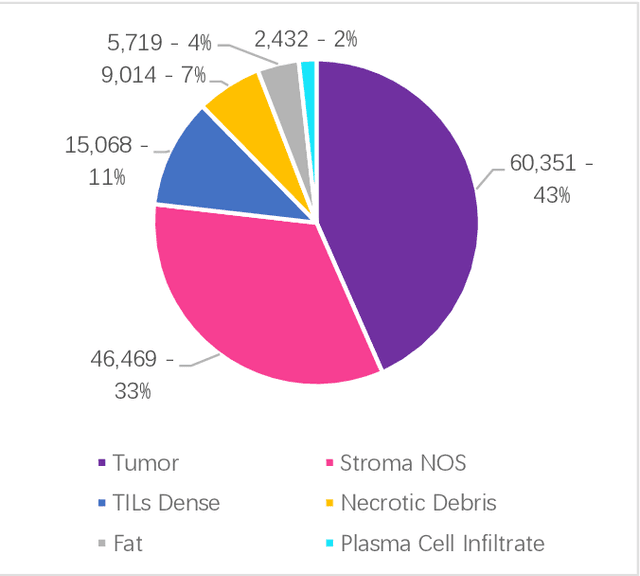
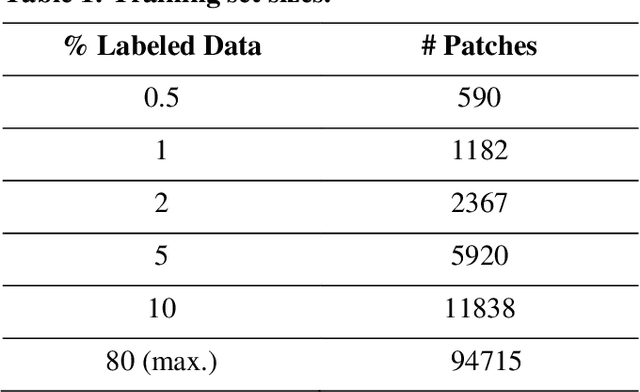
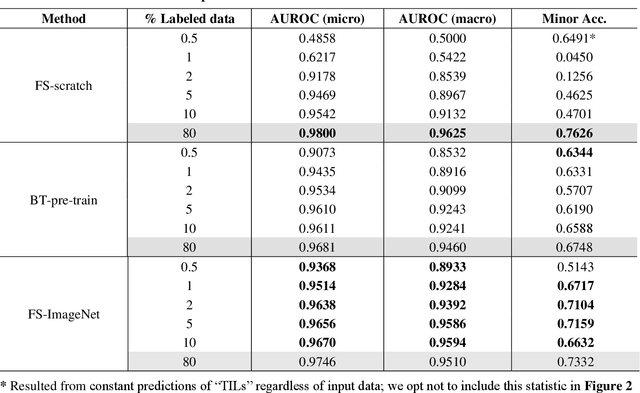
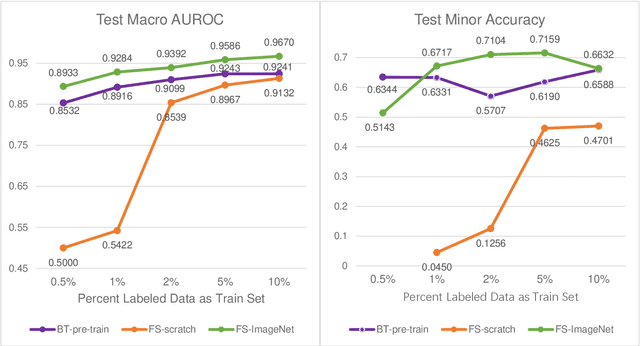
Data labeling is often the most challenging task when developing computational pathology models. Pathologist participation is necessary to generate accurate labels, and the limitations on pathologist time and demand for large, labeled datasets has led to research in areas including weakly supervised learning using patient-level labels, machine assisted annotation and active learning. In this paper we explore self-supervised learning to reduce labeling burdens in computational pathology. We explore this in the context of classification of breast cancer tissue using the Barlow Twins approach, and we compare self-supervision with alternatives like pre-trained networks in low-data scenarios. For the task explored in this paper, we find that ImageNet pre-trained networks largely outperform the self-supervised representations obtained using Barlow Twins.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge