Alvaro Fernandez-Quilez
Beyond the Signal: Medication State Effect on EEG-Based AI models for Parkinson's Disease
Mar 27, 2025Abstract:Parkinson's disease (PD) poses a growing challenge due to its increasing prevalence, complex pathology, and functional ramifications. Electroencephalography (EEG), when integrated with artificial intelligence (AI), holds promise for monitoring disease progression, identifying sub-phenotypes, and personalizing treatment strategies. However, the effect of medication state on AI model learning and generalization remains poorly understood, potentially limiting EEG-based AI models clinical applicability. This study evaluates how medication state influences the training and generalization of EEG-based AI models. Paired EEG recordings were utilized from individuals with PD in both ON- and OFF-medication states. AI models were trained on recordings from each state separately and evaluated on independent test sets representing both ON- and OFF-medication conditions. Model performance was assessed using multiple metrics, with accuracy (ACC) as the primary outcome. Statistical significance was assessed via permutation testing (p-values<0.05). Our results reveal that models trained on OFF-medication data exhibited consistent but suboptimal performance across both medication states (ACC_OFF-ON=55.3\pm8.8 and ACC_OFF-OFF=56.2\pm8.7). In contrast, models trained on ON-medication data demonstrated significantly higher performance on ON-medication recordings (ACC_ON-ON=80.7\pm7.1) but significantly reduced generalization to OFF-medication data (ACC_ON-OFF=76.0\pm7.2). Notably, models trained on ON-medication data consistently outperformed those trained on OFF-medication data within their respective states (ACC_ON-ON=80.7\pm7.1 and ACC_OFF-OFF=56.2\pm8.7). Our findings suggest that medication state significantly influences the patterns learned by AI models. Addressing this challenge is essential to enhance the robustness and clinical utility of AI models for PD characterization and management.
Reconsidering evaluation practices in modular systems: On the propagation of errors in MRI prostate cancer detection
Sep 15, 2023Abstract:Magnetic resonance imaging has evolved as a key component for prostate cancer (PCa) detection, substantially increasing the radiologist workload. Artificial intelligence (AI) systems can support radiological assessment by segmenting and classifying lesions in clinically significant (csPCa) and non-clinically significant (ncsPCa). Commonly, AI systems for PCa detection involve an automatic prostate segmentation followed by the lesion detection using the extracted prostate. However, evaluation reports are typically presented in terms of detection under the assumption of the availability of a highly accurate segmentation and an idealistic scenario, omitting the propagation of errors between modules. For that purpose, we evaluate the effect of two different segmentation networks (s1 and s2) with heterogeneous performances in the detection stage and compare it with an idealistic setting (s1:89.90+-2.23 vs 88.97+-3.06 ncsPCa, P<.001, 89.30+-4.07 and 88.12+-2.71 csPCa, P<.001). Our results depict the relevance of a holistic evaluation, accounting for all the sub-modules involved in the system.
Out-of-distribution multi-view auto-encoders for prostate cancer lesion detection
Aug 12, 2023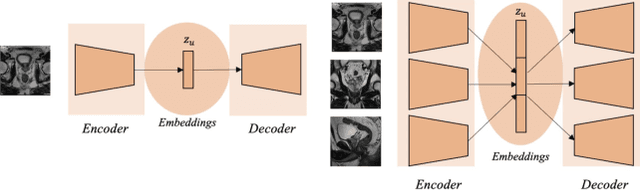
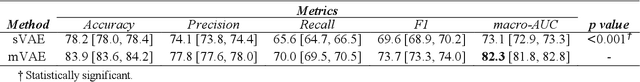
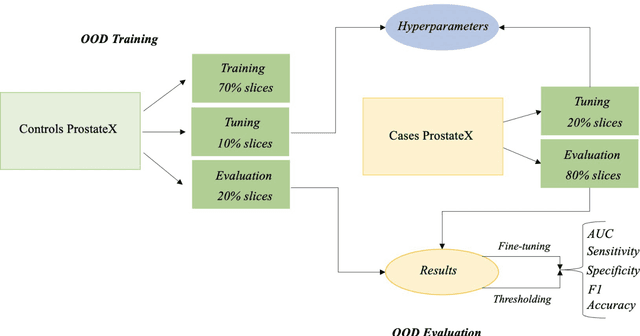
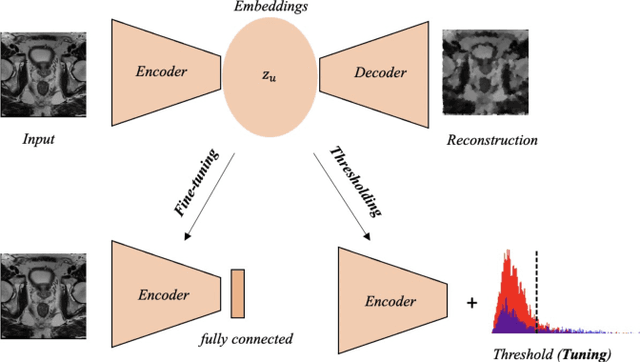
Abstract:Traditional deep learning (DL) approaches based on supervised learning paradigms require large amounts of annotated data that are rarely available in the medical domain. Unsupervised Out-of-distribution (OOD) detection is an alternative that requires less annotated data. Further, OOD applications exploit the class skewness commonly present in medical data. Magnetic resonance imaging (MRI) has proven to be useful for prostate cancer (PCa) diagnosis and management, but current DL approaches rely on T2w axial MRI, which suffers from low out-of-plane resolution. We propose a multi-stream approach to accommodate different T2w directions to improve the performance of PCa lesion detection in an OOD approach. We evaluate our approach on a publicly available data-set, obtaining better detection results in terms of AUC when compared to a single direction approach (73.1 vs 82.3). Our results show the potential of OOD approaches for PCa lesion detection based on MRI.
Leveraging multi-view data without annotations for prostate MRI segmentation: A contrastive approach
Aug 12, 2023Abstract:An accurate prostate delineation and volume characterization can support the clinical assessment of prostate cancer. A large amount of automatic prostate segmentation tools consider exclusively the axial MRI direction in spite of the availability as per acquisition protocols of multi-view data. Further, when multi-view data is exploited, manual annotations and availability at test time for all the views is commonly assumed. In this work, we explore a contrastive approach at training time to leverage multi-view data without annotations and provide flexibility at deployment time in the event of missing views. We propose a triplet encoder and single decoder network based on U-Net, tU-Net (triplet U-Net). Our proposed architecture is able to exploit non-annotated sagittal and coronal views via contrastive learning to improve the segmentation from a volumetric perspective. For that purpose, we introduce the concept of inter-view similarity in the latent space. To guide the training, we combine a dice score loss calculated with respect to the axial view and its manual annotations together with a multi-view contrastive loss. tU-Net shows statistical improvement in dice score coefficient (DSC) with respect to only axial view (91.25+-0.52% compared to 86.40+-1.50%,P<.001). Sensitivity analysis reveals the volumetric positive impact of the contrastive loss when paired with tU-Net (2.85+-1.34% compared to 3.81+-1.88%,P<.001). Further, our approach shows good external volumetric generalization in an in-house dataset when tested with multi-view data (2.76+-1.89% compared to 3.92+-3.31%,P=.002), showing the feasibility of exploiting non-annotated multi-view data through contrastive learning whilst providing flexibility at deployment in the event of missing views.
Prostate Age Gap : An MRI surrogate marker of aging for prostate cancer detection
Aug 10, 2023Abstract:Background: Prostate cancer (PC) MRI-based risk calculators are commonly based on biological (e.g. PSA), MRI markers (e.g. volume), and patient age. Whilst patient age measures the amount of years an individual has existed, biological age (BA) might better reflect the physiology of an individual. However, surrogates from prostate MRI and linkage with clinically significant PC (csPC) remain to be explored. Purpose: To obtain and evaluate Prostate Age Gap (PAG) as an MRI marker tool for csPC risk. Study type: Retrospective. Population: A total of 7243 prostate MRI slices from 468 participants who had undergone prostate biopsies. A deep learning model was trained on 3223 MRI slices cropped around the gland from 81 low-grade PC (ncsPC, Gleason score <=6) and 131 negative cases and tested on the remaining 256 participants. Assessment: Chronological age was defined as the age of the participant at the time of the visit and used to train the deep learning model to predict the age of the patient. Following, we obtained PAG, defined as the model predicted age minus the patient's chronological age. Multivariate logistic regression models were used to estimate the association through odds ratio (OR) and predictive value of PAG and compared against PSA levels and PI-RADS>=3. Statistical tests: T-test, Mann-Whitney U test, Permutation test and ROC curve analysis. Results: The multivariate adjusted model showed a significant difference in the odds of clinically significant PC (csPC, Gleason score >=7) (OR =3.78, 95% confidence interval (CI):2.32-6.16, P <.001). PAG showed a better predictive ability when compared to PI-RADS>=3 and adjusted by other risk factors, including PSA levels: AUC =0.981 vs AUC =0.704, p<.001. Conclusion: PAG was significantly associated with the risk of clinically significant PC and outperformed other well-established PC risk factors.
Assessing gender fairness in EEG-based machine learning detection of Parkinson's disease: A multi-center study
Mar 11, 2023



Abstract:As the number of automatic tools based on machine learning (ML) and resting-state electroencephalography (rs-EEG) for Parkinson's disease (PD) detection keeps growing, the assessment of possible exacerbation of health disparities by means of fairness and bias analysis becomes more relevant. Protected attributes, such as gender, play an important role in PD diagnosis development. However, analysis of sub-group populations stemming from different genders is seldom taken into consideration in ML models' development or the performance assessment for PD detection. In this work, we perform a systematic analysis of the detection ability for gender sub-groups in a multi-center setting of a previously developed ML algorithm based on power spectral density (PSD) features of rs-EEG. We find significant differences in the PD detection ability for males and females at testing time (80.5% vs. 63.7% accuracy) and significantly higher activity for a set of parietal and frontal EEG channels and frequency sub-bands for PD and non-PD males that might explain the differences in the PD detection ability for the gender sub-groups.
Machine Learning-Based Detection of Parkinson's Disease From Resting-State EEG: A Multi-Center Study
Mar 02, 2023Abstract:Resting-state EEG (rs-EEG) has been demonstrated to aid in Parkinson's disease (PD) diagnosis. In particular, the power spectral density (PSD) of low-frequency bands ({\delta} and {\theta}) and high-frequency bands ({\alpha} and \b{eta}) has been shown to be significantly different in patients with PD as compared to subjects without PD (non-PD). However, rs-EEG feature extraction and the interpretation thereof can be time-intensive and prone to examiner variability. Machine learning (ML) has the potential to automatize the analysis of rs-EEG recordings and provides a supportive tool for clinicians to ease their workload. In this work, we use rs-EEG recordings of 84 PD and 85 non-PD subjects pooled from four datasets obtained at different centers. We propose an end-to-end pipeline consisting of preprocessing, extraction of PSD features from clinically validated frequency bands, and feature selection before evaluating the classification ability of the features via ML algorithms to stratify between PD and non-PD subjects. Further, we evaluate the effect of feature harmonization, given the multi-center nature of the datasets. Our validation results show, on average, an improvement in PD detection ability (69.6% vs. 75.5% accuracy) by logistic regression when harmonizing the features and performing univariate feature selection (k = 202 features). Our final results show an average global accuracy of 72.2% with balanced accuracy results for all the centers included in the study: 60.6%, 68.7%, 77.7%, and 82.2%, respectively.
3D Masked Modelling Advances Lesion Classification in Axial T2w Prostate MRI
Dec 29, 2022Abstract:Masked Image Modelling (MIM) has been shown to be an efficient self-supervised learning (SSL) pre-training paradigm when paired with transformer architectures and in the presence of a large amount of unlabelled natural images. The combination of the difficulties in accessing and obtaining large amounts of labeled data and the availability of unlabelled data in the medical imaging domain makes MIM an interesting approach to advance deep learning (DL) applications based on 3D medical imaging data. Nevertheless, SSL and, in particular, MIM applications with medical imaging data are rather scarce and there is still uncertainty. around the potential of such a learning paradigm in the medical domain. We study MIM in the context of Prostate Cancer (PCa) lesion classification with T2 weighted (T2w) axial magnetic resonance imaging (MRI) data. In particular, we explore the effect of using MIM when coupled with convolutional neural networks (CNNs) under different conditions such as different masking strategies, obtaining better results in terms of AUC than other pre-training strategies like ImageNet weight initialization.
Self-transfer learning via patches: A prostate cancer triage approach based on bi-parametric MRI
Jul 22, 2021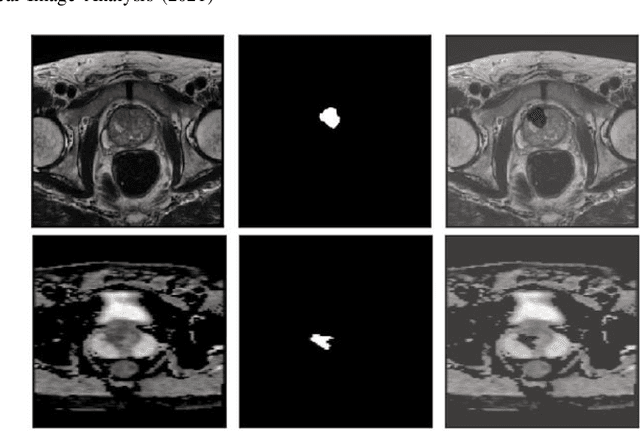
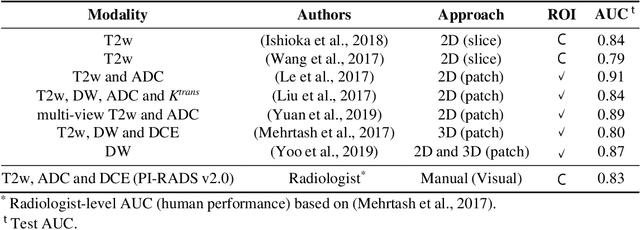
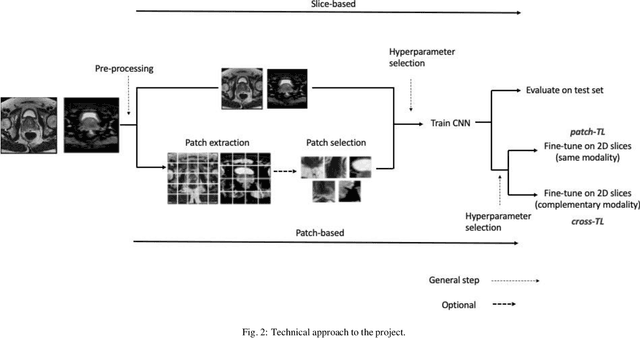
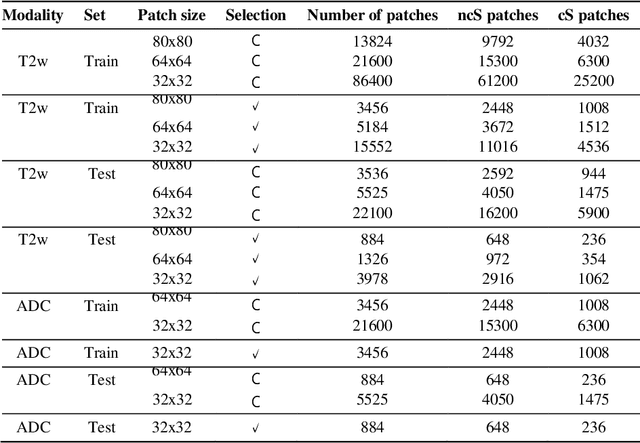
Abstract:Prostate cancer (PCa) is the second most common cancer diagnosed among men worldwide. The current PCa diagnostic pathway comes at the cost of substantial overdiagnosis, leading to unnecessary treatment and further testing. Bi-parametric magnetic resonance imaging (bp-MRI) based on apparent diffusion coefficient maps (ADC) and T2-weighted (T2w) sequences has been proposed as a triage test to differentiate between clinically significant (cS) and non-clinically significant (ncS) prostate lesions. However, analysis of the sequences relies on expertise, requires specialized training, and suffers from inter-observer variability. Deep learning (DL) techniques hold promise in tasks such as classification and detection. Nevertheless, they rely on large amounts of annotated data which is not common in the medical field. In order to palliate such issues, existing works rely on transfer learning (TL) and ImageNet pre-training, which has been proven to be sub-optimal for the medical imaging domain. In this paper, we present a patch-based pre-training strategy to distinguish between cS and ncS lesions which exploit the region of interest (ROI) of the patched source domain to efficiently train a classifier in the full-slice target domain which does not require annotations by making use of transfer learning (TL). We provide a comprehensive comparison between several CNNs architectures and different settings which are presented as a baseline. Moreover, we explore cross-domain TL which exploits both MRI modalities and improves single modality results. Finally, we show how our approaches outperform the standard approaches by a considerable margin
Improving prostate whole gland segmentation in t2-weighted MRI with synthetically generated data
Mar 27, 2021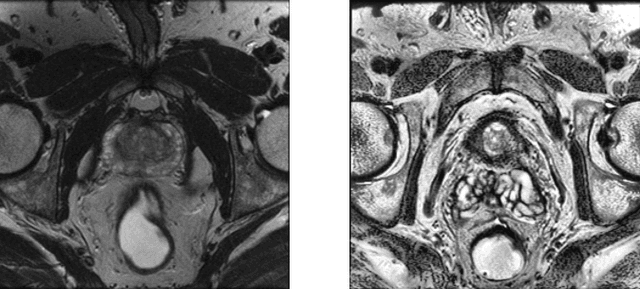

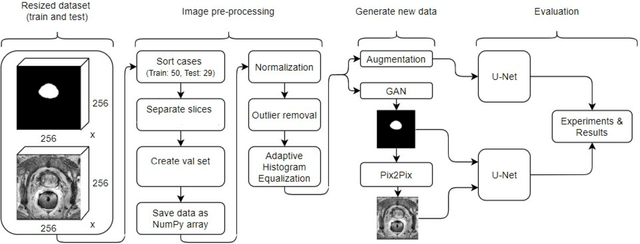

Abstract:Whole gland (WG) segmentation of the prostate plays a crucial role in detection, staging and treatment planning of prostate cancer (PCa). Despite promise shown by deep learning (DL) methods, they rely on the availability of a considerable amount of annotated data. Augmentation techniques such as translation and rotation of images present an alternative to increase data availability. Nevertheless, the amount of information provided by the transformed data is limited due to the correlation between the generated data and the original. Based on the recent success of generative adversarial networks (GAN) in producing synthetic images for other domains as well as in the medical domain, we present a pipeline to generate WG segmentation masks and synthesize T2-weighted MRI of the prostate based on a publicly available multi-center dataset. Following, we use the generated data as a form of data augmentation. Results show an improvement in the quality of the WG segmentation when compared to standard augmentation techniques.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge