Satyananda Kashyap
Modern Hopfield Networks meet Encoded Neural Representations -- Addressing Practical Considerations
Sep 24, 2024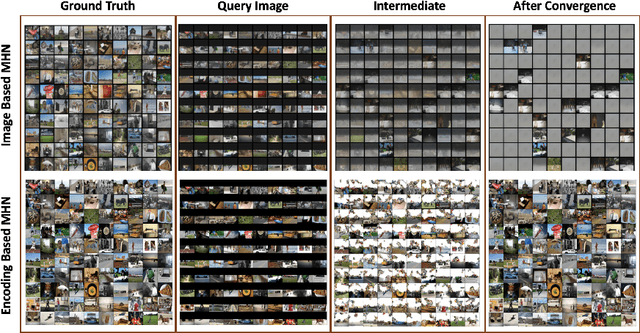
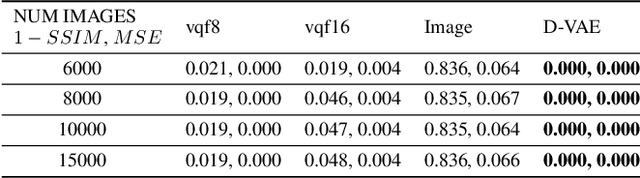
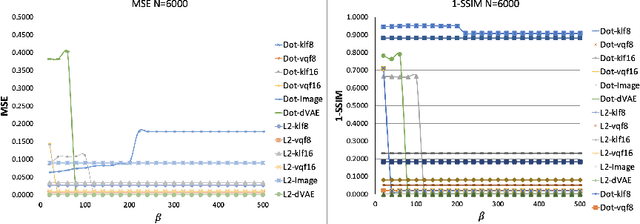
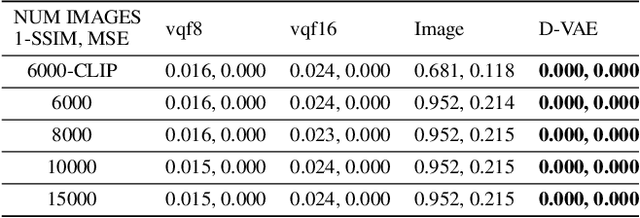
Abstract:Content-addressable memories such as Modern Hopfield Networks (MHN) have been studied as mathematical models of auto-association and storage/retrieval in the human declarative memory, yet their practical use for large-scale content storage faces challenges. Chief among them is the occurrence of meta-stable states, particularly when handling large amounts of high dimensional content. This paper introduces Hopfield Encoding Networks (HEN), a framework that integrates encoded neural representations into MHNs to improve pattern separability and reduce meta-stable states. We show that HEN can also be used for retrieval in the context of hetero association of images with natural language queries, thus removing the limitation of requiring access to partial content in the same domain. Experimental results demonstrate substantial reduction in meta-stable states and increased storage capacity while still enabling perfect recall of a significantly larger number of inputs advancing the practical utility of associative memory networks for real-world tasks.
Geo-UNet: A Geometrically Constrained Neural Framework for Clinical-Grade Lumen Segmentation in Intravascular Ultrasound
Aug 09, 2024



Abstract:Precisely estimating lumen boundaries in intravascular ultrasound (IVUS) is needed for sizing interventional stents to treat deep vein thrombosis (DVT). Unfortunately, current segmentation networks like the UNet lack the precision needed for clinical adoption in IVUS workflows. This arises due to the difficulty of automatically learning accurate lumen contour from limited training data while accounting for the radial geometry of IVUS imaging. We propose the Geo-UNet framework to address these issues via a design informed by the geometry of the lumen contour segmentation task. We first convert the input data and segmentation targets from Cartesian to polar coordinates. Starting from a convUNet feature extractor, we propose a two-task setup, one for conventional pixel-wise labeling and the other for single boundary lumen-contour localization. We directly combine the two predictions by passing the predicted lumen contour through a new activation (named CDFeLU) to filter out spurious pixel-wise predictions. Our unified loss function carefully balances area-based, distance-based, and contour-based penalties to provide near clinical-grade generalization in unseen patient data. We also introduce a lightweight, inference-time technique to enhance segmentation smoothness. The efficacy of our framework on a venous IVUS dataset is shown against state-of-the-art models.
Towards Automatic Prediction of Outcome in Treatment of Cerebral Aneurysms
Nov 18, 2022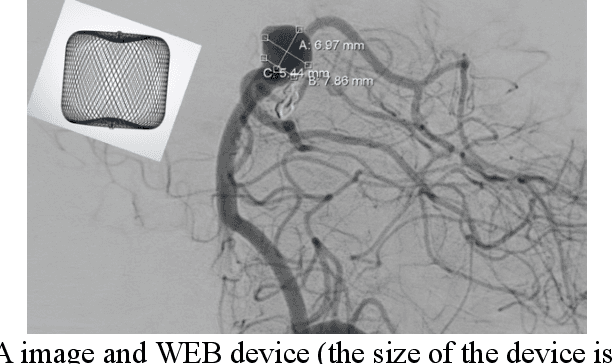
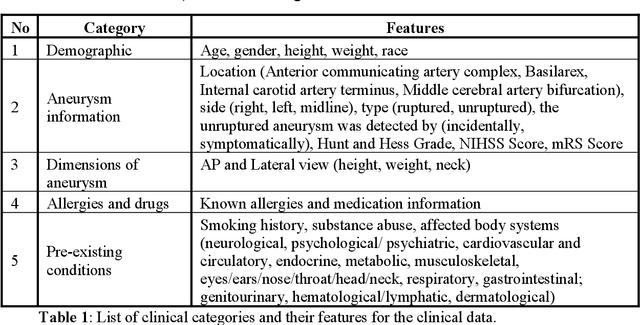
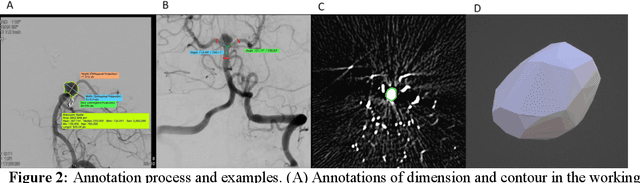
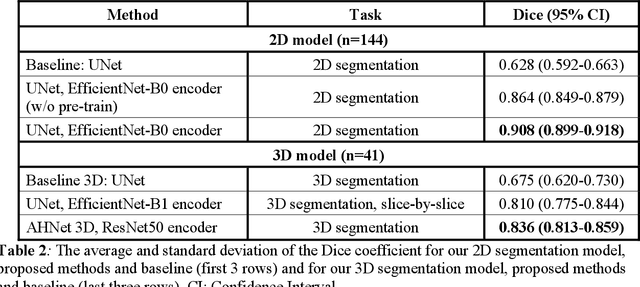
Abstract:Intrasaccular flow disruptors treat cerebral aneurysms by diverting the blood flow from the aneurysm sac. Residual flow into the sac after the intervention is a failure that could be due to the use of an undersized device, or to vascular anatomy and clinical condition of the patient. We report a machine learning model based on over 100 clinical and imaging features that predict the outcome of wide-neck bifurcation aneurysm treatment with an intravascular embolization device. We combine clinical features with a diverse set of common and novel imaging measurements within a random forest model. We also develop neural network segmentation algorithms in 2D and 3D to contour the sac in angiographic images and automatically calculate the imaging features. These deliver 90% overlap with manual contouring in 2D and 83% in 3D. Our predictive model classifies complete vs. partial occlusion outcomes with an accuracy of 75.31%, and weighted F1-score of 0.74.
* 10 pages
MedPerf: Open Benchmarking Platform for Medical Artificial Intelligence using Federated Evaluation
Oct 08, 2021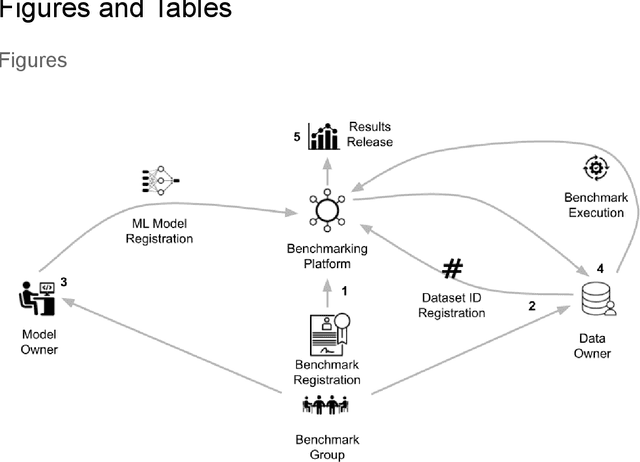
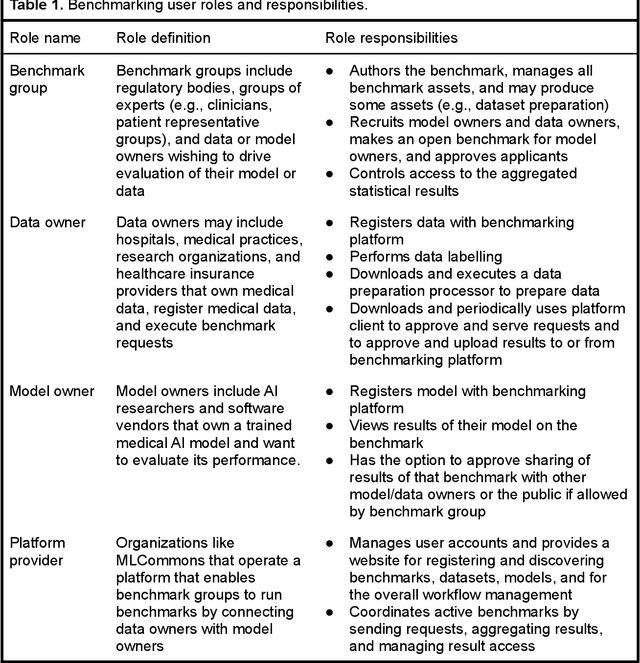
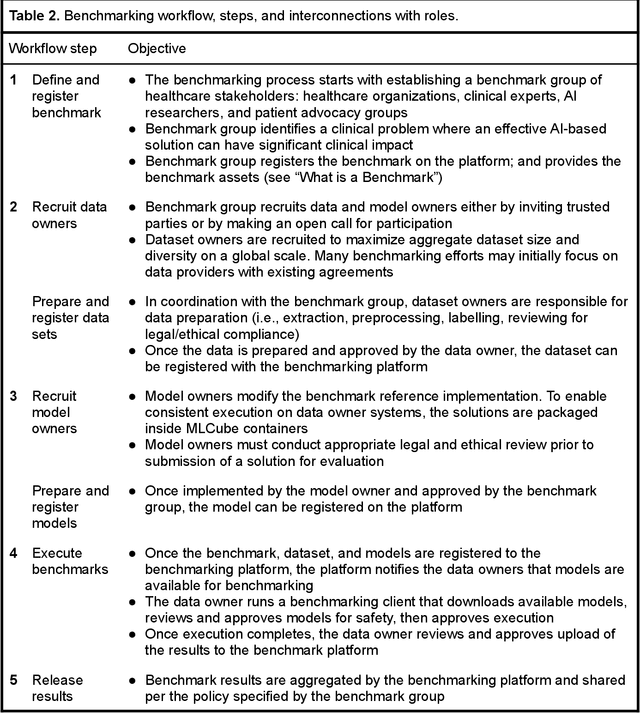
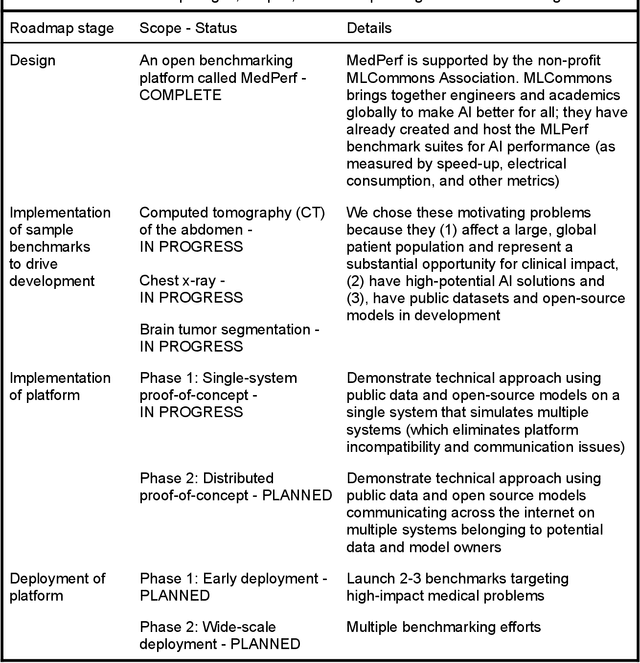
Abstract:Medical AI has tremendous potential to advance healthcare by supporting the evidence-based practice of medicine, personalizing patient treatment, reducing costs, and improving provider and patient experience. We argue that unlocking this potential requires a systematic way to measure the performance of medical AI models on large-scale heterogeneous data. To meet this need, we are building MedPerf, an open framework for benchmarking machine learning in the medical domain. MedPerf will enable federated evaluation in which models are securely distributed to different facilities for evaluation, thereby empowering healthcare organizations to assess and verify the performance of AI models in an efficient and human-supervised process, while prioritizing privacy. We describe the current challenges healthcare and AI communities face, the need for an open platform, the design philosophy of MedPerf, its current implementation status, and our roadmap. We call for researchers and organizations to join us in creating the MedPerf open benchmarking platform.
Basis Scaling and Double Pruning for Efficient Transfer Learning
Aug 06, 2021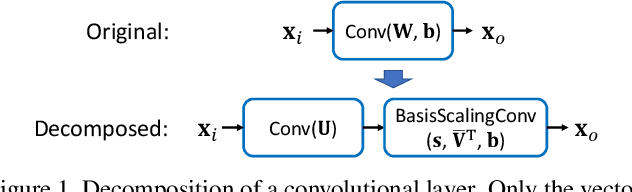
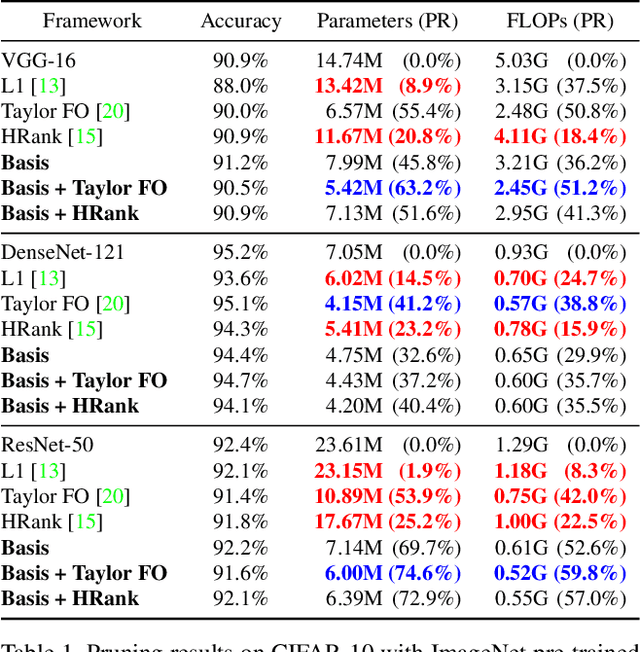
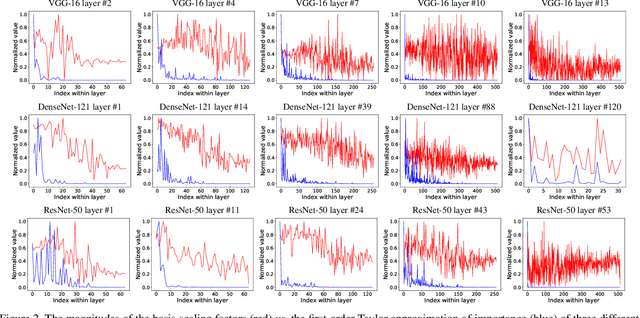
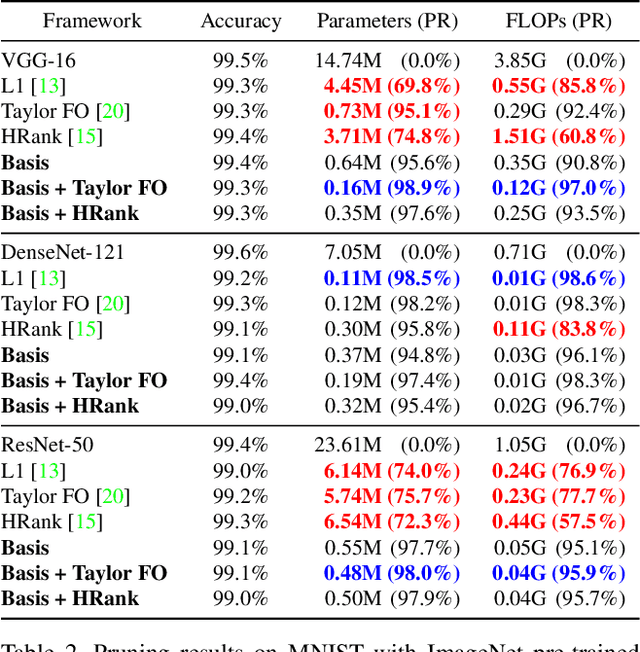
Abstract:Transfer learning allows the reuse of deep learning features on new datasets with limited data. However, the resulting models could be unnecessarily large and thus inefficient. Although network pruning can be applied to improve inference efficiency, existing algorithms usually require fine-tuning and may not be suitable for small datasets. In this paper, we propose an algorithm that transforms the convolutional weights into the subspaces of orthonormal bases where a model is pruned. Using singular value decomposition, we decompose a convolutional layer into two layers: a convolutional layer with the orthonormal basis vectors as the filters, and a layer that we name "BasisScalingConv", which is responsible for rescaling the features and transforming them back to the original space. As the filters in each transformed layer are linearly independent with known relative importance, pruning can be more effective and stable, and fine tuning individual weights is unnecessary. Furthermore, as the numbers of input and output channels of the original convolutional layer remain unchanged, basis pruning is applicable to virtually all network architectures. Basis pruning can also be combined with existing pruning algorithms for double pruning to further increase the pruning capability. With less than 1% reduction in the classification accuracy, we can achieve pruning ratios up to 98.9% in parameters and 98.6% in FLOPs.
Chest ImaGenome Dataset for Clinical Reasoning
Jul 31, 2021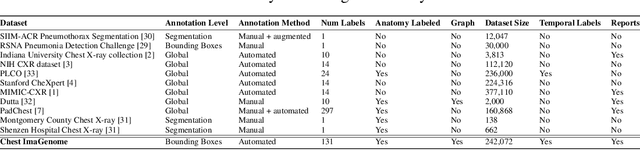
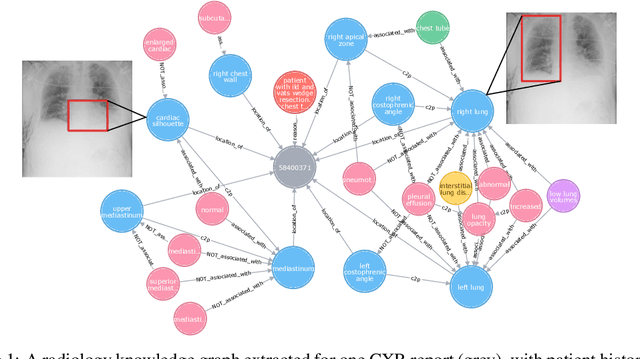
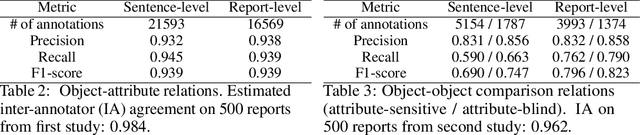
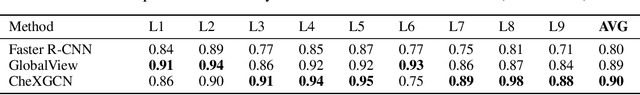
Abstract:Despite the progress in automatic detection of radiologic findings from chest X-ray (CXR) images in recent years, a quantitative evaluation of the explainability of these models is hampered by the lack of locally labeled datasets for different findings. With the exception of a few expert-labeled small-scale datasets for specific findings, such as pneumonia and pneumothorax, most of the CXR deep learning models to date are trained on global "weak" labels extracted from text reports, or trained via a joint image and unstructured text learning strategy. Inspired by the Visual Genome effort in the computer vision community, we constructed the first Chest ImaGenome dataset with a scene graph data structure to describe $242,072$ images. Local annotations are automatically produced using a joint rule-based natural language processing (NLP) and atlas-based bounding box detection pipeline. Through a radiologist constructed CXR ontology, the annotations for each CXR are connected as an anatomy-centered scene graph, useful for image-level reasoning and multimodal fusion applications. Overall, we provide: i) $1,256$ combinations of relation annotations between $29$ CXR anatomical locations (objects with bounding box coordinates) and their attributes, structured as a scene graph per image, ii) over $670,000$ localized comparison relations (for improved, worsened, or no change) between the anatomical locations across sequential exams, as well as ii) a manually annotated gold standard scene graph dataset from $500$ unique patients.
Channel Scaling: A Scale-and-Select Approach for Transfer Learning
Mar 22, 2021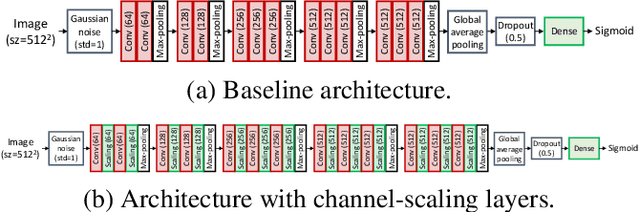
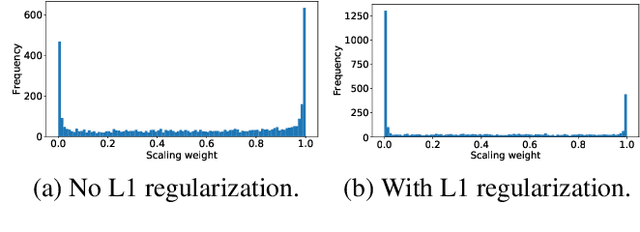
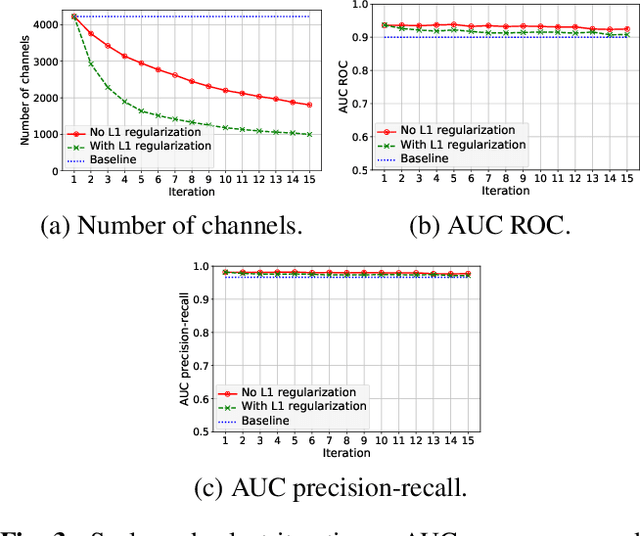
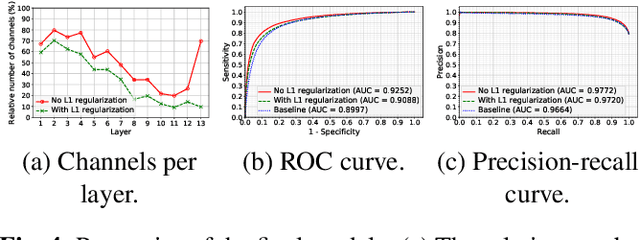
Abstract:Transfer learning with pre-trained neural networks is a common strategy for training classifiers in medical image analysis. Without proper channel selections, this often results in unnecessarily large models that hinder deployment and explainability. In this paper, we propose a novel approach to efficiently build small and well performing networks by introducing the channel-scaling layers. A channel-scaling layer is attached to each frozen convolutional layer, with the trainable scaling weights inferring the importance of the corresponding feature channels. Unlike the fine-tuning approaches, we maintain the weights of the original channels and large datasets are not required. By imposing L1 regularization and thresholding on the scaling weights, this framework iteratively removes unnecessary feature channels from a pre-trained model. Using an ImageNet pre-trained VGG16 model, we demonstrate the capabilities of the proposed framework on classifying opacity from chest X-ray images. The results show that we can reduce the number of parameters by 95% while delivering a superior performance.
Creation and Validation of a Chest X-Ray Dataset with Eye-tracking and Report Dictation for AI Development
Oct 08, 2020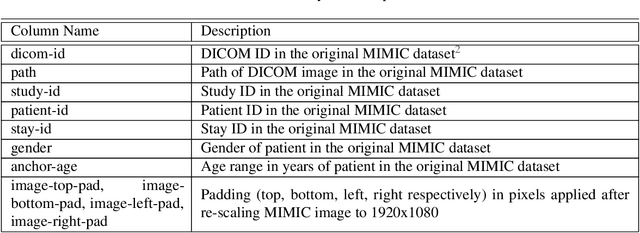
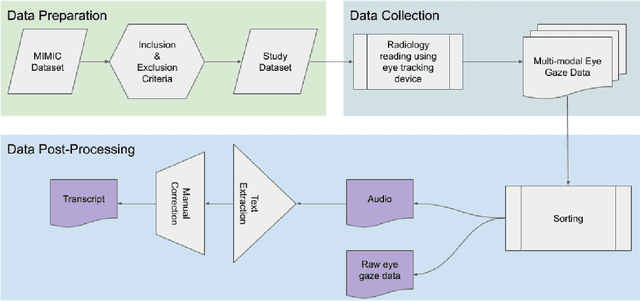
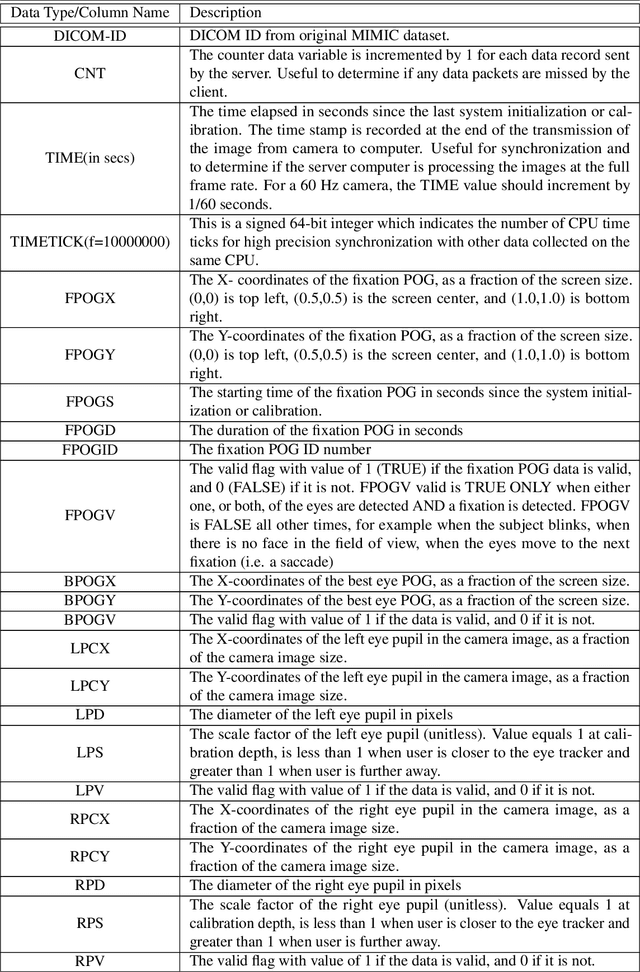
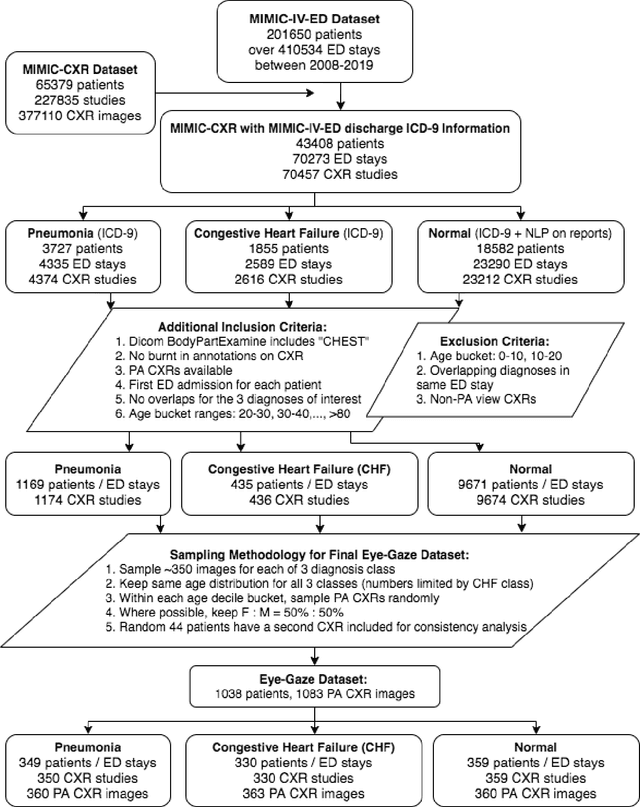
Abstract:We developed a rich dataset of Chest X-Ray (CXR) images to assist investigators in artificial intelligence. The data were collected using an eye tracking system while a radiologist reviewed and reported on 1,083 CXR images. The dataset contains the following aligned data: CXR image, transcribed radiology report text, radiologist's dictation audio and eye gaze coordinates data. We hope this dataset can contribute to various areas of research particularly towards explainable and multimodal deep learning / machine learning methods. Furthermore, investigators in disease classification and localization, automated radiology report generation, and human-machine interaction can benefit from these data. We report deep learning experiments that utilize the attention maps produced by eye gaze dataset to show the potential utility of this data.
Learning Invariant Feature Representation to Improve Generalization across Chest X-ray Datasets
Aug 04, 2020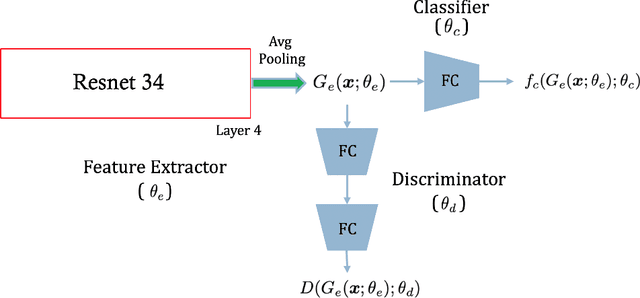
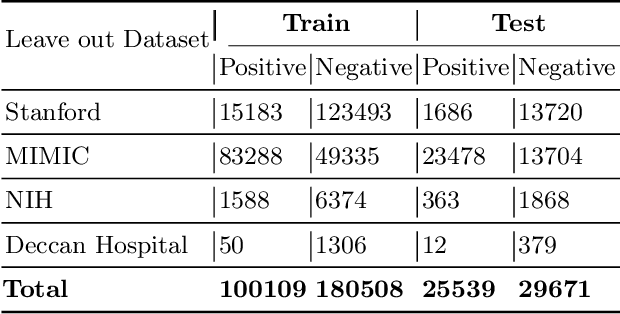

Abstract:Chest radiography is the most common medical image examination for screening and diagnosis in hospitals. Automatic interpretation of chest X-rays at the level of an entry-level radiologist can greatly benefit work prioritization and assist in analyzing a larger population. Subsequently, several datasets and deep learning-based solutions have been proposed to identify diseases based on chest X-ray images. However, these methods are shown to be vulnerable to shift in the source of data: a deep learning model performing well when tested on the same dataset as training data, starts to perform poorly when it is tested on a dataset from a different source. In this work, we address this challenge of generalization to a new source by forcing the network to learn a source-invariant representation. By employing an adversarial training strategy, we show that a network can be forced to learn a source-invariant representation. Through pneumonia-classification experiments on multi-source chest X-ray datasets, we show that this algorithm helps in improving classification accuracy on a new source of X-ray dataset.
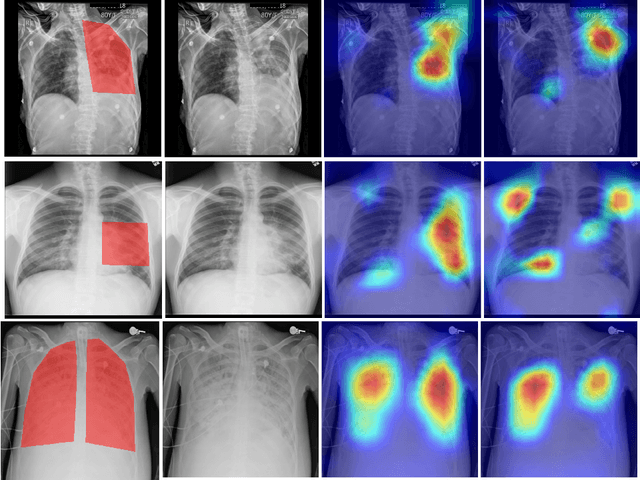
Looking in the Right place for Anomalies: Explainable AI through Automatic Location Learning
Aug 02, 2020
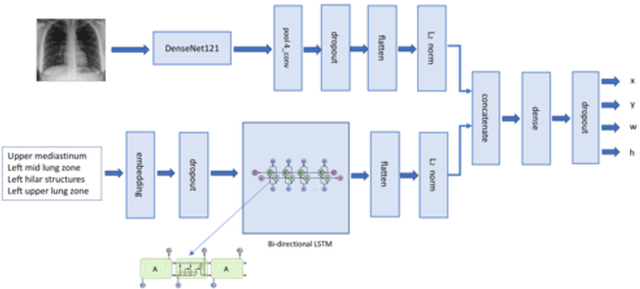
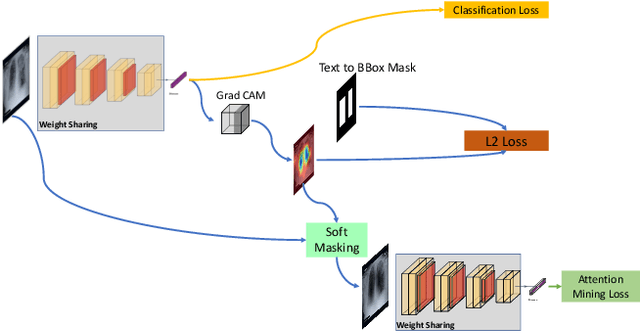
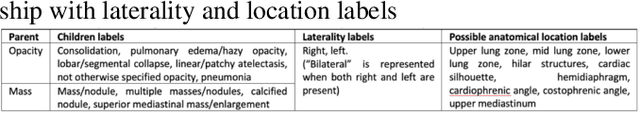
Abstract:Deep learning has now become the de facto approach to the recognition of anomalies in medical imaging. Their 'black box' way of classifying medical images into anomaly labels poses problems for their acceptance, particularly with clinicians. Current explainable AI methods offer justifications through visualizations such as heat maps but cannot guarantee that the network is focusing on the relevant image region fully containing the anomaly. In this paper, we develop an approach to explainable AI in which the anomaly is assured to be overlapping the expected location when present. This is made possible by automatically extracting location-specific labels from textual reports and learning the association of expected locations to labels using a hybrid combination of Bi-Directional Long Short-Term Memory Recurrent Neural Networks (Bi-LSTM) and DenseNet-121. Use of this expected location to bias the subsequent attention-guided inference network based on ResNet101 results in the isolation of the anomaly at the expected location when present. The method is evaluated on a large chest X-ray dataset.
* 5 pages, Paper presented as a poster at the International Symposium on Biomedical Imaging, 2020, Paper Number 655
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge