Derek H. Oakley
Improving Neuropathological Reconstruction Fidelity via AI Slice Imputation
Jan 31, 2026Abstract:Neuropathological analyses benefit from spatially precise volumetric reconstructions that enhance anatomical delineation and improve morphometric accuracy. Our prior work has shown the feasibility of reconstructing 3D brain volumes from 2D dissection photographs. However these outputs sometimes exhibit coarse, overly smooth reconstructions of structures, especially under high anisotropy (i.e., reconstructions from thick slabs). Here, we introduce a computationally efficient super-resolution step that imputes slices to generate anatomically consistent isotropic volumes from anisotropic 3D reconstructions of dissection photographs. By training on domain-randomized synthetic data, we ensure that our method generalizes across dissection protocols and remains robust to large slab thicknesses. The imputed volumes yield improved automated segmentations, achieving higher Dice scores, particularly in cortical and white matter regions. Validation on surface reconstruction and atlas registration tasks demonstrates more accurate cortical surfaces and MRI registration. By enhancing the resolution and anatomical fidelity of photograph-based reconstructions, our approach strengthens the bridge between neuropathology and neuroimaging. Our method is publicly available at https://surfer.nmr.mgh.harvard.edu/fswiki/mri_3d_photo_recon
Reference-Free 3D Reconstruction of Brain Dissection Photographs with Machine Learning
Mar 13, 2025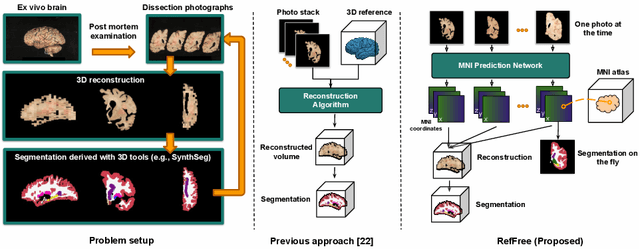
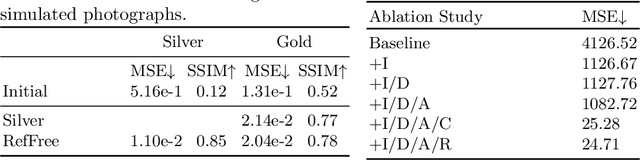
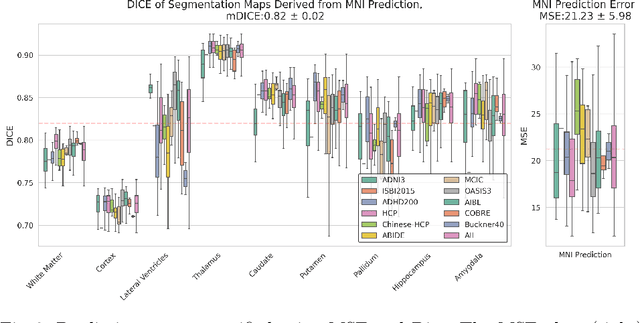
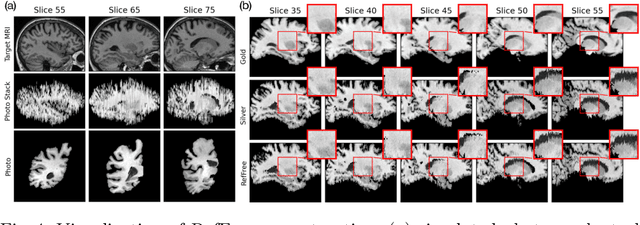
Abstract:Correlation of neuropathology with MRI has the potential to transfer microscopic signatures of pathology to invivo scans. Recently, a classical registration method has been proposed, to build these correlations from 3D reconstructed stacks of dissection photographs, which are routinely taken at brain banks. These photographs bypass the need for exvivo MRI, which is not widely accessible. However, this method requires a full stack of brain slabs and a reference mask (e.g., acquired with a surface scanner), which severely limits the applicability of the technique. Here we propose RefFree, a dissection photograph reconstruction method without external reference. RefFree is a learning approach that estimates the 3D coordinates in the atlas space for every pixel in every photograph; simple least-squares fitting can then be used to compute the 3D reconstruction. As a by-product, RefFree also produces an atlas-based segmentation of the reconstructed stack. RefFree is trained on synthetic photographs generated from digitally sliced 3D MRI data, with randomized appearance for enhanced generalization ability. Experiments on simulated and real data show that RefFree achieves performance comparable to the baseline method without an explicit reference while also enabling reconstruction of partial stacks. Our code is available at https://github.com/lintian-a/reffree.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge