Zhengyi Lu
Explainable Pathomics Feature Visualization via Correlation-aware Conditional Feature Editing
Feb 05, 2026Abstract:Pathomics is a recent approach that offers rich quantitative features beyond what black-box deep learning can provide, supporting more reproducible and explainable biomarkers in digital pathology. However, many derived features (e.g., "second-order moment") remain difficult to interpret, especially across different clinical contexts, which limits their practical adoption. Conditional diffusion models show promise for explainability through feature editing, but they typically assume feature independence**--**an assumption violated by intrinsically correlated pathomics features. Consequently, editing one feature while fixing others can push the model off the biological manifold and produce unrealistic artifacts. To address this, we propose a Manifold-Aware Diffusion (MAD) framework for controllable and biologically plausible cell nuclei editing. Unlike existing approaches, our method regularizes feature trajectories within a disentangled latent space learned by a variational auto-encoder (VAE). This ensures that manipulating a target feature automatically adjusts correlated attributes to remain within the learned distribution of real cells. These optimized features then guide a conditional diffusion model to synthesize high-fidelity images. Experiments demonstrate that our approach is able to navigate the manifold of pathomics features when editing those features. The proposed method outperforms baseline methods in conditional feature editing while preserving structural coherence.
Local-Global Multimodal Contrastive Learning for Molecular Property Prediction
Jan 30, 2026Abstract:Accurate molecular property prediction requires integrating complementary information from molecular structure and chemical semantics. In this work, we propose LGM-CL, a local-global multimodal contrastive learning framework that jointly models molecular graphs and textual representations derived from SMILES and chemistry-aware augmented texts. Local functional group information and global molecular topology are captured using AttentiveFP and Graph Transformer encoders, respectively, and aligned through self-supervised contrastive learning. In addition, chemically enriched textual descriptions are contrasted with original SMILES to incorporate physicochemical semantics in a task-agnostic manner. During fine-tuning, molecular fingerprints are further integrated via Dual Cross-attention multimodal fusion. Extensive experiments on MoleculeNet benchmarks demonstrate that LGM-CL achieves consistent and competitive performance across both classification and regression tasks, validating the effectiveness of unified local-global and multimodal representation learning.
AdaFuse: Adaptive Multimodal Fusion for Lung Cancer Risk Prediction via Reinforcement Learning
Jan 30, 2026Abstract:Multimodal fusion has emerged as a promising paradigm for disease diagnosis and prognosis, integrating complementary information from heterogeneous data sources such as medical images, clinical records, and radiology reports. However, existing fusion methods process all available modalities through the network, either treating them equally or learning to assign different contribution weights, leaving a fundamental question unaddressed: for a given patient, should certain modalities be used at all? We present AdaFuse, an adaptive multimodal fusion framework that leverages reinforcement learning (RL) to learn patient-specific modality selection and fusion strategies for lung cancer risk prediction. AdaFuse formulates multimodal fusion as a sequential decision process, where the policy network iteratively decides whether to incorporate an additional modality or proceed to prediction based on the information already acquired. This sequential formulation enables the model to condition each selection on previously observed modalities and terminate early when sufficient information is available, rather than committing to a fixed subset upfront. We evaluate AdaFuse on the National Lung Screening Trial (NLST) dataset. Experimental results demonstrate that AdaFuse achieves the highest AUC (0.762) compared to the best single-modality baseline (0.732), the best fixed fusion strategy (0.759), and adaptive baselines including DynMM (0.754) and MoE (0.742), while using fewer FLOPs than all triple-modality methods. Our work demonstrates the potential of reinforcement learning for personalized multimodal fusion in medical imaging, representing a shift from uniform fusion strategies toward adaptive diagnostic pipelines that learn when to consult additional modalities and when existing information suffices for accurate prediction.
MASC: Metal-Aware Sampling and Correction via Reinforcement Learning for Accelerated MRI
Jan 30, 2026Abstract:Metal implants in MRI cause severe artifacts that degrade image quality and hinder clinical diagnosis. Traditional approaches address metal artifact reduction (MAR) and accelerated MRI acquisition as separate problems. We propose MASC, a unified reinforcement learning framework that jointly optimizes metal-aware k-space sampling and artifact correction for accelerated MRI. To enable supervised training, we construct a paired MRI dataset using physics-based simulation, generating k-space data and reconstructions for phantoms with and without metal implants. This paired dataset provides simulated 3D MRI scans with and without metal implants, where each metal-corrupted sample has an exactly matched clean reference, enabling direct supervision for both artifact reduction and acquisition policy learning. We formulate active MRI acquisition as a sequential decision-making problem, where an artifact-aware Proximal Policy Optimization (PPO) agent learns to select k-space phase-encoding lines under a limited acquisition budget. The agent operates on undersampled reconstructions processed through a U-Net-based MAR network, learning patterns that maximize reconstruction quality. We further propose an end-to-end training scheme where the acquisition policy learns to select k-space lines that best support artifact removal while the MAR network simultaneously adapts to the resulting undersampling patterns. Experiments demonstrate that MASC's learned policies outperform conventional sampling strategies, and end-to-end training improves performance compared to using a frozen pre-trained MAR network, validating the benefit of joint optimization. Cross-dataset experiments on FastMRI with physics-based artifact simulation further confirm generalization to realistic clinical MRI data. The code and models of MASC have been made publicly available: https://github.com/hrlblab/masc
SCR2-ST: Combine Single Cell with Spatial Transcriptomics for Efficient Active Sampling via Reinforcement Learning
Dec 15, 2025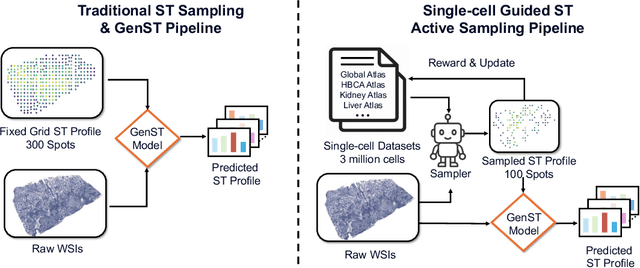
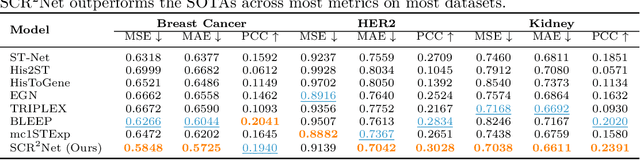
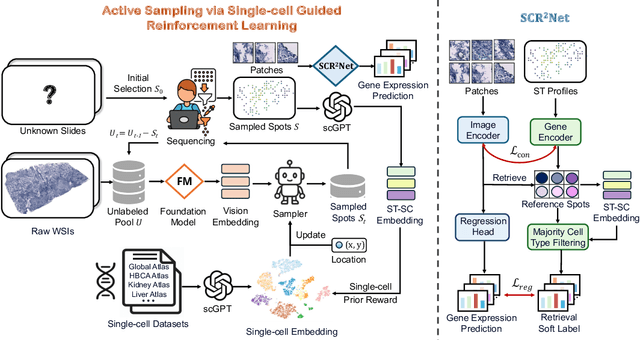
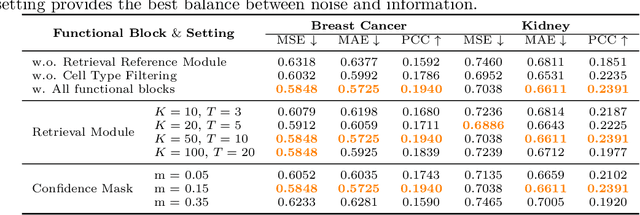
Abstract:Spatial transcriptomics (ST) is an emerging technology that enables researchers to investigate the molecular relationships underlying tissue morphology. However, acquiring ST data remains prohibitively expensive, and traditional fixed-grid sampling strategies lead to redundant measurements of morphologically similar or biologically uninformative regions, thus resulting in scarce data that constrain current methods. The well-established single-cell sequencing field, however, could provide rich biological data as an effective auxiliary source to mitigate this limitation. To bridge these gaps, we introduce SCR2-ST, a unified framework that leverages single-cell prior knowledge to guide efficient data acquisition and accurate expression prediction. SCR2-ST integrates a single-cell guided reinforcement learning-based (SCRL) active sampling and a hybrid regression-retrieval prediction network SCR2Net. SCRL combines single-cell foundation model embeddings with spatial density information to construct biologically grounded reward signals, enabling selective acquisition of informative tissue regions under constrained sequencing budgets. SCR2Net then leverages the actively sampled data through a hybrid architecture combining regression-based modeling with retrieval-augmented inference, where a majority cell-type filtering mechanism suppresses noisy matches and retrieved expression profiles serve as soft labels for auxiliary supervision. We evaluated SCR2-ST on three public ST datasets, demonstrating SOTA performance in both sampling efficiency and prediction accuracy, particularly under low-budget scenarios. Code is publicly available at: https://github.com/hrlblab/SCR2ST
CASC-AI: Consensus-aware Self-corrective AI Agents for Noise Cell Segmentation
Feb 11, 2025Abstract:Multi-class cell segmentation in high-resolution gigapixel whole slide images (WSI) is crucial for various clinical applications. However, training such models typically requires labor-intensive, pixel-wise annotations by domain experts. Recent efforts have democratized this process by involving lay annotators without medical expertise. However, conventional non-agent-based approaches struggle to handle annotation noise adaptively, as they lack mechanisms to mitigate false positives (FP) and false negatives (FN) at both the image-feature and pixel levels. In this paper, we propose a consensus-aware self-corrective AI agent that leverages the Consensus Matrix to guide its learning process. The Consensus Matrix defines regions where both the AI and annotators agree on cell and non-cell annotations, which are prioritized with stronger supervision. Conversely, areas of disagreement are adaptively weighted based on their feature similarity to high-confidence agreement regions, with more similar regions receiving greater attention. Additionally, contrastive learning is employed to separate features of noisy regions from those of reliable agreement regions by maximizing their dissimilarity. This paradigm enables the AI to iteratively refine noisy labels, enhancing its robustness. Validated on one real-world lay-annotated cell dataset and two simulated noisy datasets, our method demonstrates improved segmentation performance, effectively correcting FP and FN errors and showcasing its potential for training robust models on noisy datasets. The official implementation and cell annotations are publicly available at https://github.com/ddrrnn123/CASC-AI.
Fast-RF-Shimming: Accelerate RF Shimming in 7T MRI using Deep Learning
Jan 21, 2025Abstract:Ultrahigh field (UHF) Magnetic Resonance Imaging (MRI) provides a high signal-to-noise ratio (SNR), enabling exceptional spatial resolution for clinical diagnostics and research. However, higher fields introduce challenges such as transmit radiofrequency (RF) field inhomogeneities, which result in uneven flip angles and image intensity artifacts. These artifacts degrade image quality and limit clinical adoption. Traditional RF shimming methods, including Magnitude Least Squares (MLS) optimization, mitigate RF field inhomogeneity but are time-intensive and often require the presence of the patient. Recent machine learning methods, such as RF Shim Prediction by Iteratively Projected Ridge Regression and other deep learning architectures, offer alternative approaches but face challenges such as extensive training requirements, limited complexity, and practical data constraints. This paper introduces a holistic learning-based framework called Fast RF Shimming, which achieves a 5000-fold speedup compared to MLS methods. First, random-initialized Adaptive Moment Estimation (Adam) derives reference shimming weights from multichannel RF fields. Next, a Residual Network (ResNet) maps RF fields to shimming outputs while incorporating a confidence parameter into the loss function. Finally, a Non-uniformity Field Detector (NFD) identifies extreme non-uniform outcomes. Comparative evaluations demonstrate significant improvements in both speed and predictive accuracy. The proposed pipeline also supports potential extensions, such as the integration of anatomical priors or multi-echo data, to enhance the robustness of RF field correction. This approach offers a faster and more efficient solution to RF shimming challenges in UHF MRI.
Optimizing Transmit Field Inhomogeneity of Parallel RF Transmit Design in 7T MRI using Deep Learning
Aug 21, 2024Abstract:Ultrahigh field (UHF) Magnetic Resonance Imaging (MRI) provides a higher signal-to-noise ratio and, thereby, higher spatial resolution. However, UHF MRI introduces challenges such as transmit radiofrequency (RF) field (B1+) inhomogeneities, leading to uneven flip angles and image intensity anomalies. These issues can significantly degrade imaging quality and its medical applications. This study addresses B1+ field homogeneity through a novel deep learning-based strategy. Traditional methods like Magnitude Least Squares (MLS) optimization have been effective but are time-consuming and dependent on the patient's presence. Recent machine learning approaches, such as RF Shim Prediction by Iteratively Projected Ridge Regression and deep learning frameworks, have shown promise but face limitations like extensive training times and oversimplified architectures. We propose a two-step deep learning strategy. First, we obtain the desired reference RF shimming weights from multi-channel B1+ fields using random-initialized Adaptive Moment Estimation. Then, we employ Residual Networks (ResNets) to train a model that maps B1+ fields to target RF shimming outputs. Our approach does not rely on pre-calculated reference optimizations for the testing process and efficiently learns residual functions. Comparative studies with traditional MLS optimization demonstrate our method's advantages in terms of speed and accuracy. The proposed strategy achieves a faster and more efficient RF shimming design, significantly improving imaging quality at UHF. This advancement holds potential for broader applications in medical imaging and diagnostics.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge