Asim Iqbal
Governance at the Edge of Architecture: Regulating NeuroAI and Neuromorphic Systems
Feb 02, 2026Abstract:Current AI governance frameworks, including regulatory benchmarks for accuracy, latency, and energy efficiency, are built for static, centrally trained artificial neural networks on von Neumann hardware. NeuroAI systems, embodied in neuromorphic hardware and implemented via spiking neural networks, break these assumptions. This paper examines the limitations of current AI governance frameworks for NeuroAI, arguing that assurance and audit methods must co-evolve with these architectures, aligning traditional regulatory metrics with the physics, learning dynamics, and embodied efficiency of brain-inspired computation to enable technically grounded assurance.
NeuGen: Amplifying the 'Neural' in Neural Radiance Fields for Domain Generalization
May 11, 2025Abstract:Neural Radiance Fields (NeRF) have significantly advanced the field of novel view synthesis, yet their generalization across diverse scenes and conditions remains challenging. Addressing this, we propose the integration of a novel brain-inspired normalization technique Neural Generalization (NeuGen) into leading NeRF architectures which include MVSNeRF and GeoNeRF. NeuGen extracts the domain-invariant features, thereby enhancing the models' generalization capabilities. It can be seamlessly integrated into NeRF architectures and cultivates a comprehensive feature set that significantly improves accuracy and robustness in image rendering. Through this integration, NeuGen shows improved performance on benchmarks on diverse datasets across state-of-the-art NeRF architectures, enabling them to generalize better across varied scenes. Our comprehensive evaluations, both quantitative and qualitative, confirm that our approach not only surpasses existing models in generalizability but also markedly improves rendering quality. Our work exemplifies the potential of merging neuroscientific principles with deep learning frameworks, setting a new precedent for enhanced generalizability and efficiency in novel view synthesis. A demo of our study is available at https://neugennerf.github.io.
Mice to Machines: Neural Representations from Visual Cortex for Domain Generalization
May 11, 2025Abstract:The mouse is one of the most studied animal models in the field of systems neuroscience. Understanding the generalized patterns and decoding the neural representations that are evoked by the diverse range of natural scene stimuli in the mouse visual cortex is one of the key quests in computational vision. In recent years, significant parallels have been drawn between the primate visual cortex and hierarchical deep neural networks. However, their generalized efficacy in understanding mouse vision has been limited. In this study, we investigate the functional alignment between the mouse visual cortex and deep learning models for object classification tasks. We first introduce a generalized representational learning strategy that uncovers a striking resemblance between the functional mapping of the mouse visual cortex and high-performing deep learning models on both top-down (population-level) and bottom-up (single cell-level) scenarios. Next, this representational similarity across the two systems is further enhanced by the addition of Neural Response Normalization (NeuRN) layer, inspired by the activation profile of excitatory and inhibitory neurons in the visual cortex. To test the performance effect of NeuRN on real-world tasks, we integrate it into deep learning models and observe significant improvements in their robustness against data shifts in domain generalization tasks. Our work proposes a novel framework for comparing the functional architecture of the mouse visual cortex with deep learning models. Our findings carry broad implications for the development of advanced AI models that draw inspiration from the mouse visual cortex, suggesting that these models serve as valuable tools for studying the neural representations of the mouse visual cortex and, as a result, enhancing their performance on real-world tasks.
* 12 pages, 8 figures, 1 table
NeuRN: Neuro-inspired Domain Generalization for Image Classification
May 11, 2025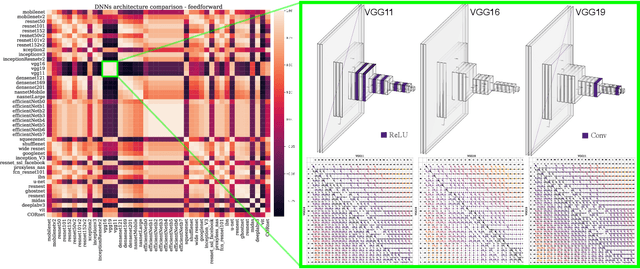
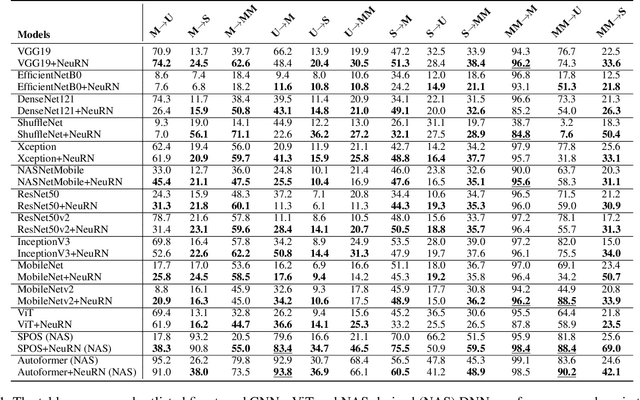
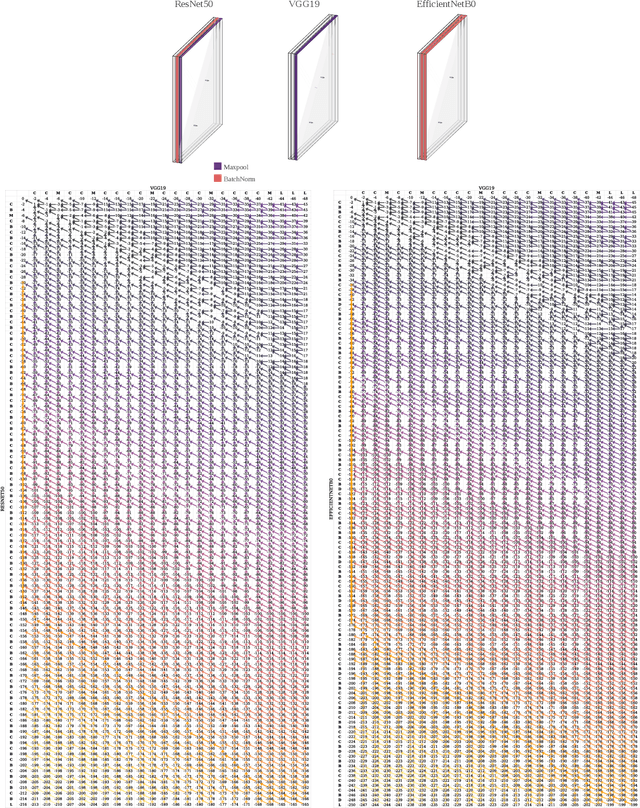
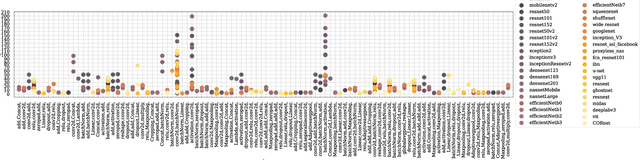
Abstract:Domain generalization in image classification is a crucial challenge, with models often failing to generalize well across unseen datasets. We address this issue by introducing a neuro-inspired Neural Response Normalization (NeuRN) layer which draws inspiration from neurons in the mammalian visual cortex, which aims to enhance the performance of deep learning architectures on unseen target domains by training deep learning models on a source domain. The performance of these models is considered as a baseline and then compared against models integrated with NeuRN on image classification tasks. We perform experiments across a range of deep learning architectures, including ones derived from Neural Architecture Search and Vision Transformer. Additionally, in order to shortlist models for our experiment from amongst the vast range of deep neural networks available which have shown promising results, we also propose a novel method that uses the Needleman-Wunsch algorithm to compute similarity between deep learning architectures. Our results demonstrate the effectiveness of NeuRN by showing improvement against baseline in cross-domain image classification tasks. Our framework attempts to establish a foundation for future neuro-inspired deep learning models.
* 14 pages, 7 figures, 1 table
NeuReg: Domain-invariant 3D Image Registration on Human and Mouse Brains
Nov 09, 2024Abstract:Medical brain imaging relies heavily on image registration to accurately curate structural boundaries of brain features for various healthcare applications. Deep learning models have shown remarkable performance in image registration in recent years. Still, they often struggle to handle the diversity of 3D brain volumes, challenged by their structural and contrastive variations and their imaging domains. In this work, we present NeuReg, a Neuro-inspired 3D image registration architecture with the feature of domain invariance. NeuReg generates domain-agnostic representations of imaging features and incorporates a shifting window-based Swin Transformer block as the encoder. This enables our model to capture the variations across brain imaging modalities and species. We demonstrate a new benchmark in multi-domain publicly available datasets comprising human and mouse 3D brain volumes. Extensive experiments reveal that our model (NeuReg) outperforms the existing baseline deep learning-based image registration models and provides a high-performance boost on cross-domain datasets, where models are trained on 'source-only' domain and tested on completely 'unseen' target domains. Our work establishes a new state-of-the-art for domain-agnostic 3D brain image registration, underpinned by Neuro-inspired Transformer-based architecture.
AnimalFormer: Multimodal Vision Framework for Behavior-based Precision Livestock Farming
Jun 14, 2024Abstract:We introduce a multimodal vision framework for precision livestock farming, harnessing the power of GroundingDINO, HQSAM, and ViTPose models. This integrated suite enables comprehensive behavioral analytics from video data without invasive animal tagging. GroundingDINO generates accurate bounding boxes around livestock, while HQSAM segments individual animals within these boxes. ViTPose estimates key body points, facilitating posture and movement analysis. Demonstrated on a sheep dataset with grazing, running, sitting, standing, and walking activities, our framework extracts invaluable insights: activity and grazing patterns, interaction dynamics, and detailed postural evaluations. Applicable across species and video resolutions, this framework revolutionizes non-invasive livestock monitoring for activity detection, counting, health assessments, and posture analyses. It empowers data-driven farm management, optimizing animal welfare and productivity through AI-powered behavioral understanding.
A comparison of automatic multi-tissue segmentation methods of the human fetal brain using the FeTA Dataset
Oct 29, 2020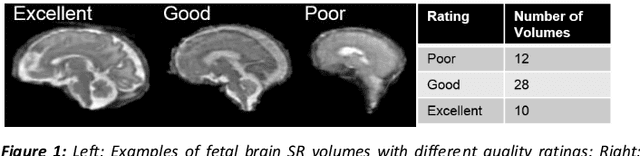
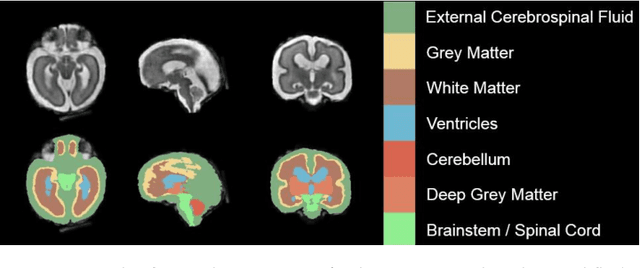
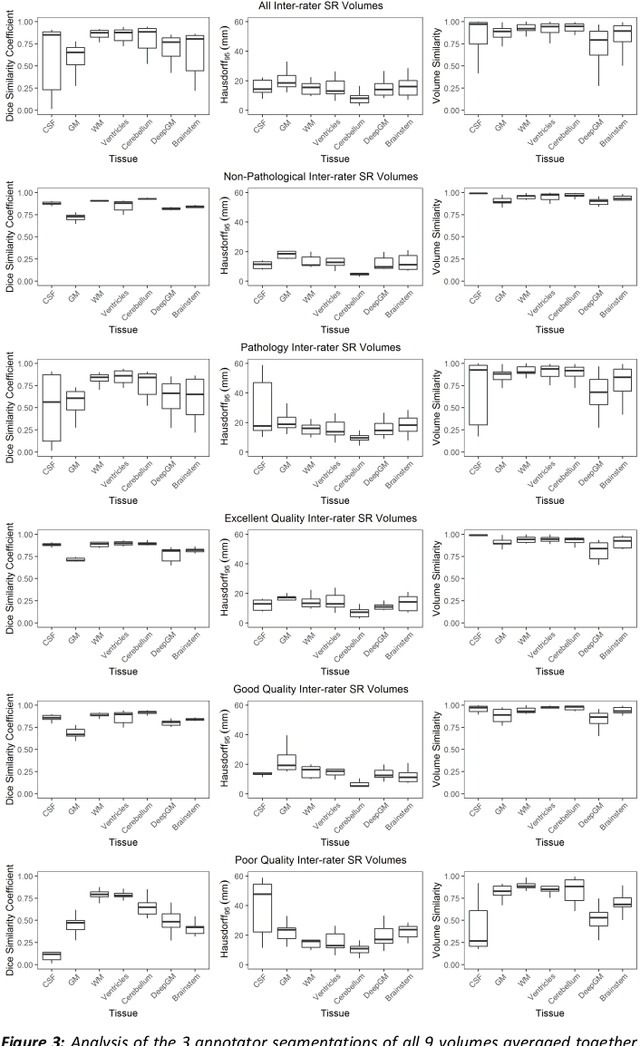
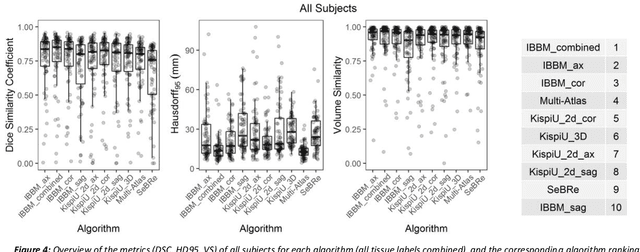
Abstract:It is critical to quantitatively analyse the developing human fetal brain in order to fully understand neurodevelopment in both normal fetuses and those with congenital disorders. To facilitate this analysis, automatic multi-tissue fetal brain segmentation algorithms are needed, which in turn requires open databases of segmented fetal brains. Here we introduce a publicly available database of 50 manually segmented pathological and non-pathological fetal magnetic resonance brain volume reconstructions across a range of gestational ages (20 to 33 weeks) into 7 different tissue categories (external cerebrospinal fluid, grey matter, white matter, ventricles, cerebellum, deep grey matter, brainstem/spinal cord). In addition, we quantitatively evaluate the accuracy of several automatic multi-tissue segmentation algorithms of the developing human fetal brain. Four research groups participated, submitting a total of 10 algorithms, demonstrating the benefits the database for the development of automatic algorithms.
Exploring Intensity Invariance in Deep Neural Networks for Brain Image Registration
Sep 21, 2020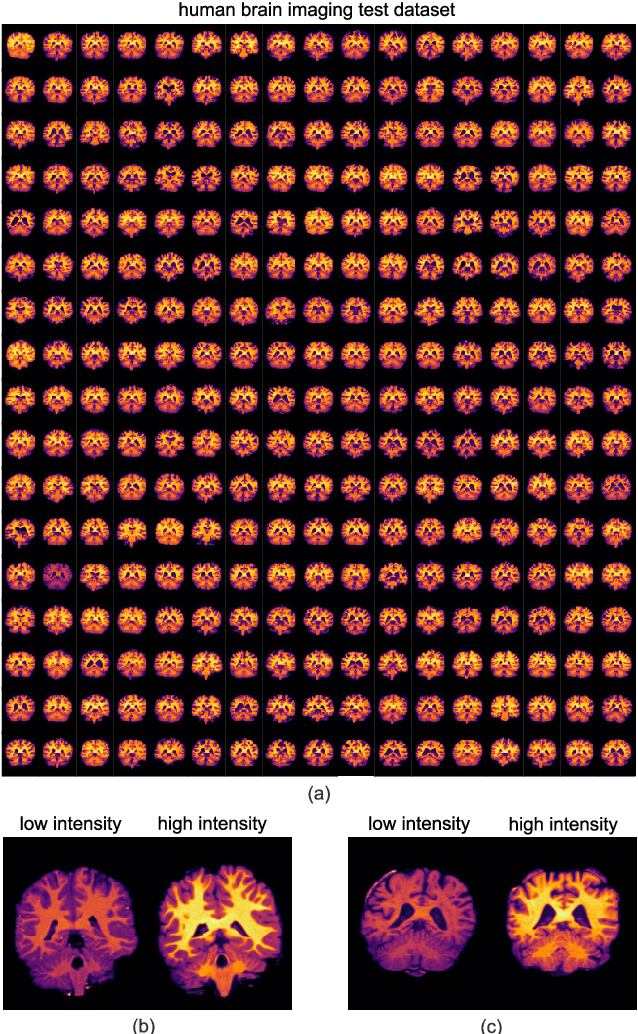
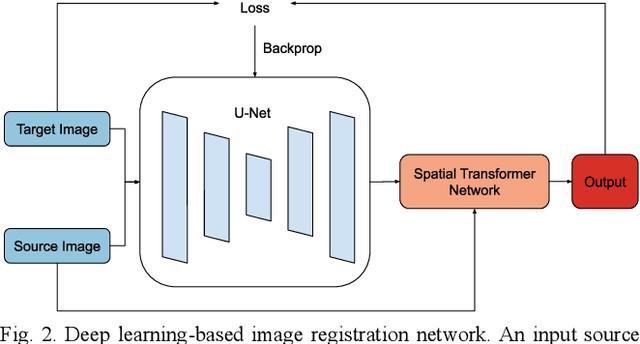
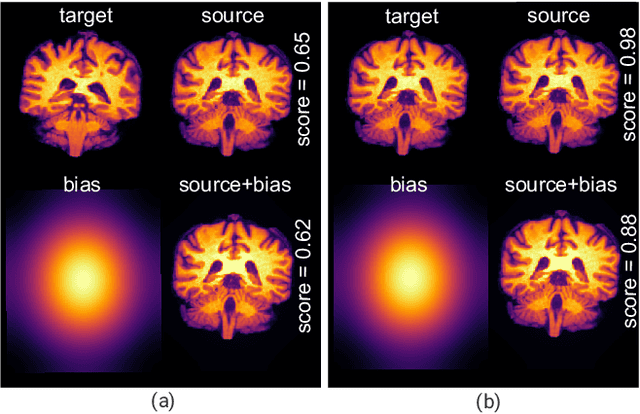
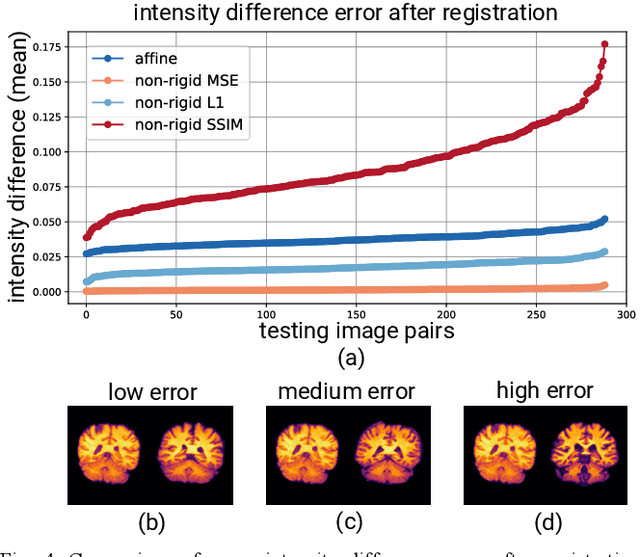
Abstract:Image registration is a widely-used technique in analysing large scale datasets that are captured through various imaging modalities and techniques in biomedical imaging such as MRI, X-Rays, etc. These datasets are typically collected from various sites and under different imaging protocols using a variety of scanners. Such heterogeneity in the data collection process causes inhomogeneity or variation in intensity (brightness) and noise distribution. These variations play a detrimental role in the performance of image registration, segmentation and detection algorithms. Classical image registration methods are computationally expensive but are able to handle these artifacts relatively better. However, deep learning-based techniques are shown to be computationally efficient for automated brain registration but are sensitive to the intensity variations. In this study, we investigate the effect of variation in intensity distribution among input image pairs for deep learning-based image registration methods. We find a performance degradation of these models when brain image pairs with different intensity distribution are presented even with similar structures. To overcome this limitation, we incorporate a structural similarity-based loss function in a deep neural network and test its performance on the validation split separated before training as well as on a completely unseen new dataset. We report that the deep learning models trained with structure similarity-based loss seems to perform better for both datasets. This investigation highlights a possible performance limiting factor in deep learning-based registration models and suggests a potential solution to incorporate the intensity distribution variation in the input image pairs. Our code and models are available at https://github.com/hassaanmahmood/DeepIntense.
Decoding Neural Responses in Mouse Visual Cortex through a Deep Neural Network
Oct 26, 2019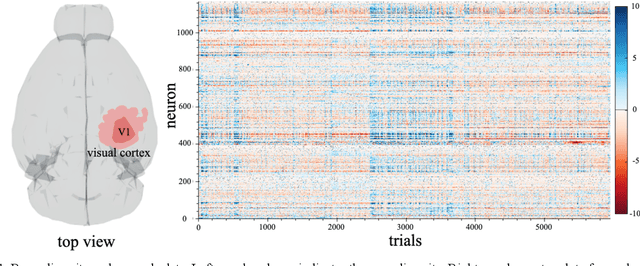
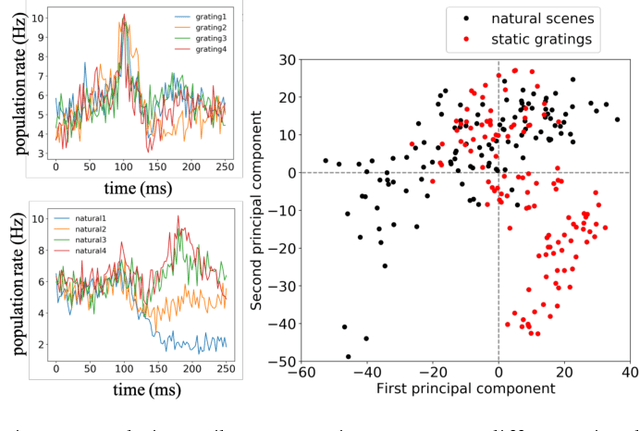
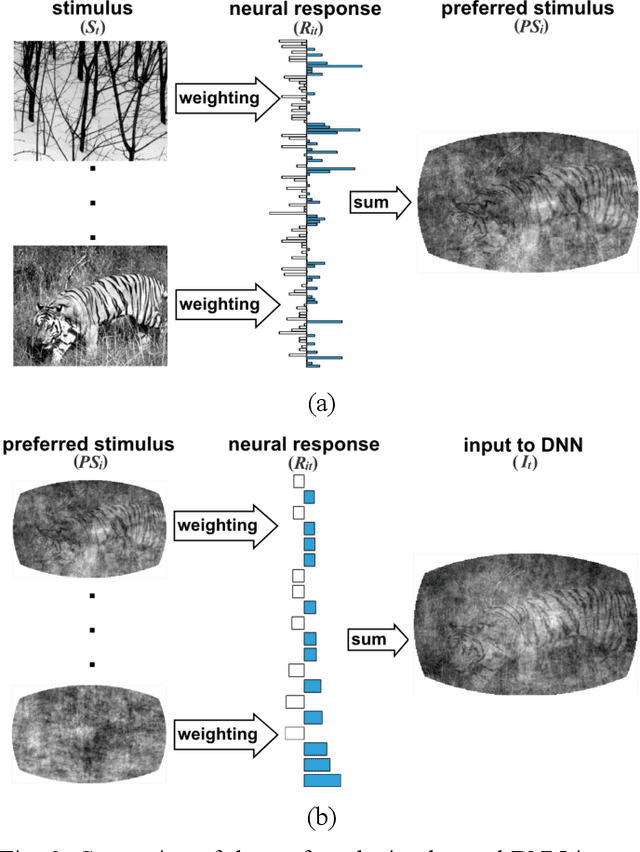
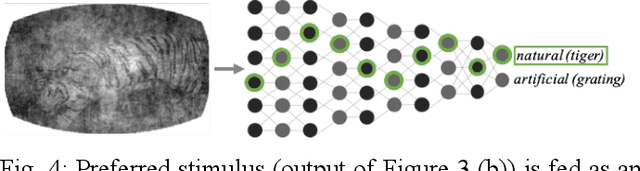
Abstract:Finding a code to unravel the population of neural responses that leads to a distinct animal behavior has been a long-standing question in the field of neuroscience. With the recent advances in machine learning, it is shown that the hierarchically Deep Neural Networks (DNNs) perform optimally in decoding unique features out of complex datasets. In this study, we utilize the power of a DNN to explore the computational principles in the mammalian brain by exploiting the Neuropixel data from Allen Brain Institute. We decode the neural responses from mouse visual cortex to predict the presented stimuli to the animal for natural (bear, trees, cheetah, etc.) and artificial (drifted gratings, orientated bars, etc.) classes. Our results indicate that neurons in mouse visual cortex encode the features of natural and artificial objects in a distinct manner, and such neural code is consistent across animals. We investigate this by applying transfer learning to train a DNN on the neural responses of a single animal and test its generalized performance across multiple animals. Within a single animal, DNN is able to decode the neural responses with as much as 100% classification accuracy. Across animals, this accuracy is reduced to 91%. This study demonstrates the potential of utilizing the DNN models as a computational framework to understand the neural coding principles in the mammalian brain.
Developing Brain Atlas through Deep Learning
Jul 10, 2018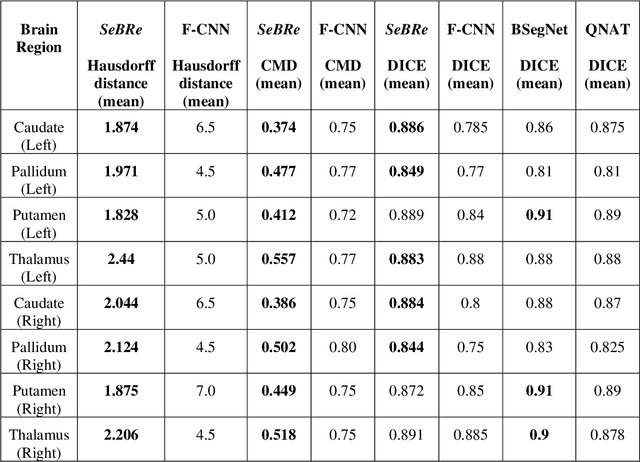
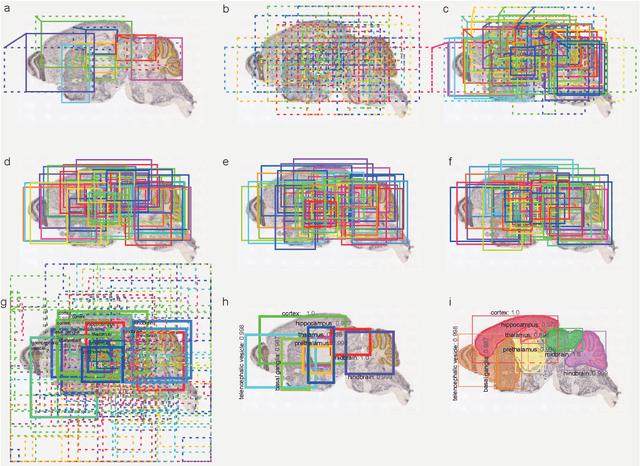
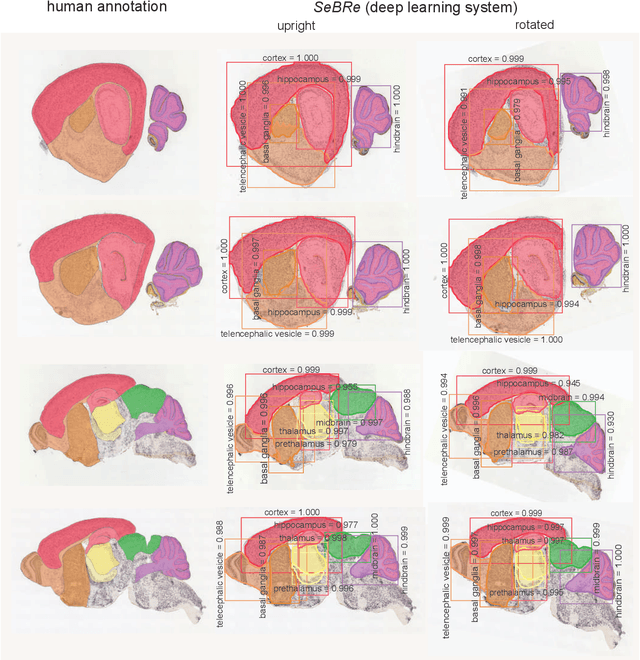
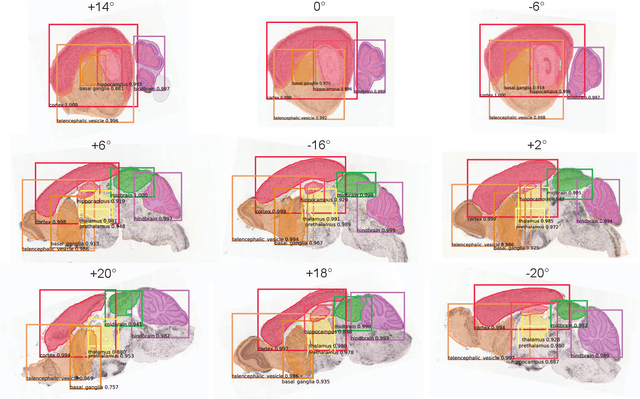
Abstract:To uncover the organizational principles governing the human brain, neuroscientists are in need of developing high-throughput methods that can explore the structure and function of distinct brain regions using animal models. The first step towards this goal is to accurately register the regions of interest in a mouse brain, against a standard reference atlas, with minimum human supervision. The second step is to scale this approach to different animal ages, so as to also allow insights into normal and pathological brain development and aging. We introduce here a fully automated convolutional neural network-based method (SeBRe) for registration through Segmenting Brain Regions of interest in mice at different ages. We demonstrate the validity of our method on different mouse brain post-natal (P) developmental time points, across a range of neuronal markers. Our method outperforms the existing brain registration methods, and provides the minimum mean squared error (MSE) score on a mouse brain dataset. We propose that our deep learning-based registration method can (i) accelerate brain-wide exploration of region-specific changes in brain development and (ii) replace the existing complex brain registration methodology, by simply segmenting brain regions of interest for high-throughput brain-wide analysis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge