Theofanis Karayannis
A comparison of automatic multi-tissue segmentation methods of the human fetal brain using the FeTA Dataset
Oct 29, 2020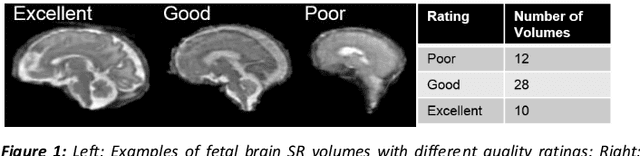
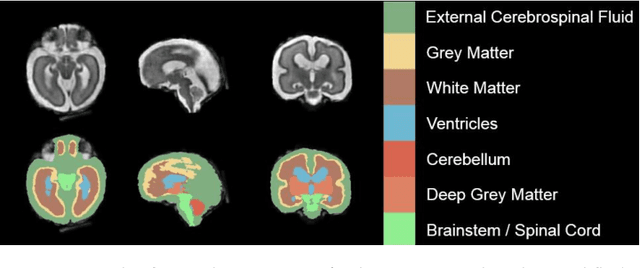
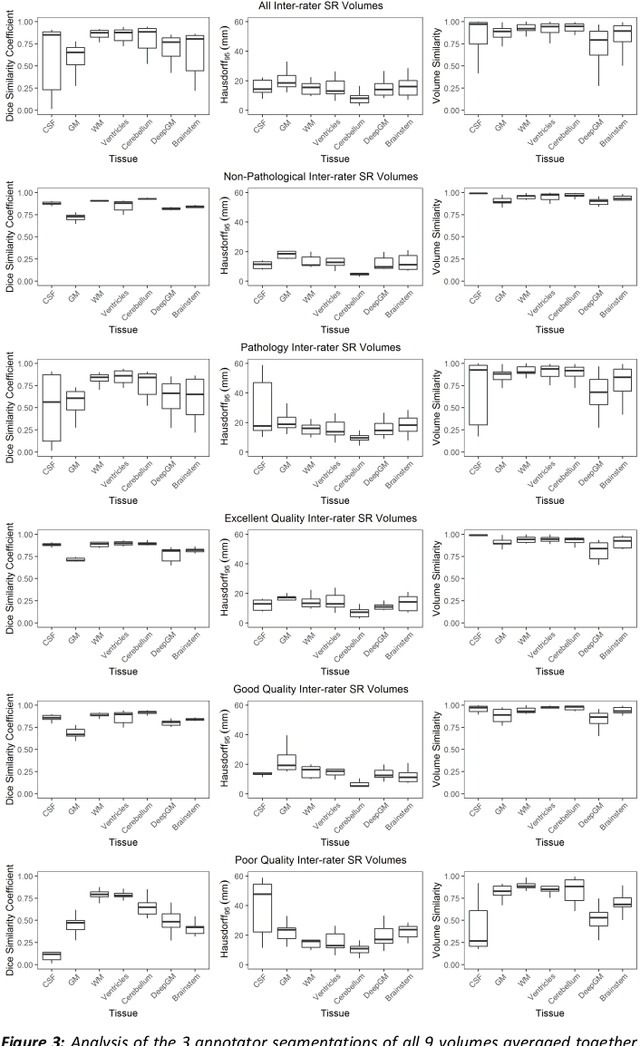
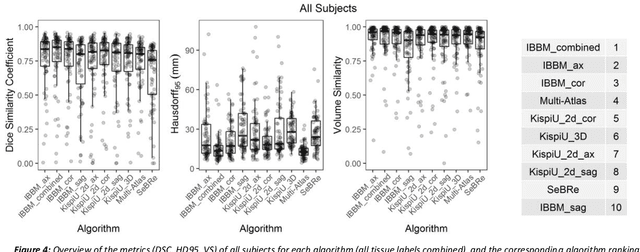
Abstract:It is critical to quantitatively analyse the developing human fetal brain in order to fully understand neurodevelopment in both normal fetuses and those with congenital disorders. To facilitate this analysis, automatic multi-tissue fetal brain segmentation algorithms are needed, which in turn requires open databases of segmented fetal brains. Here we introduce a publicly available database of 50 manually segmented pathological and non-pathological fetal magnetic resonance brain volume reconstructions across a range of gestational ages (20 to 33 weeks) into 7 different tissue categories (external cerebrospinal fluid, grey matter, white matter, ventricles, cerebellum, deep grey matter, brainstem/spinal cord). In addition, we quantitatively evaluate the accuracy of several automatic multi-tissue segmentation algorithms of the developing human fetal brain. Four research groups participated, submitting a total of 10 algorithms, demonstrating the benefits the database for the development of automatic algorithms.
Developing Brain Atlas through Deep Learning
Jul 10, 2018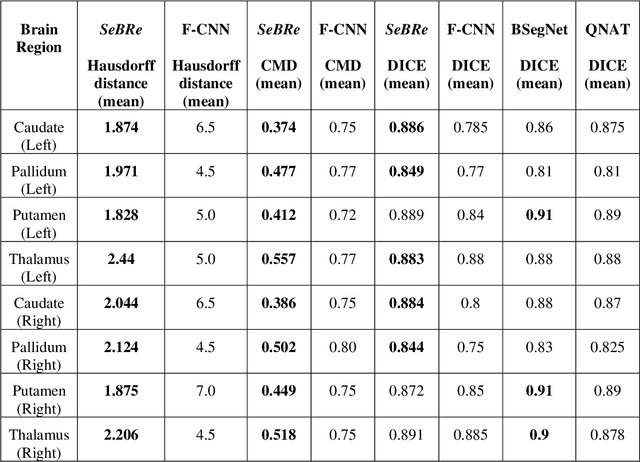
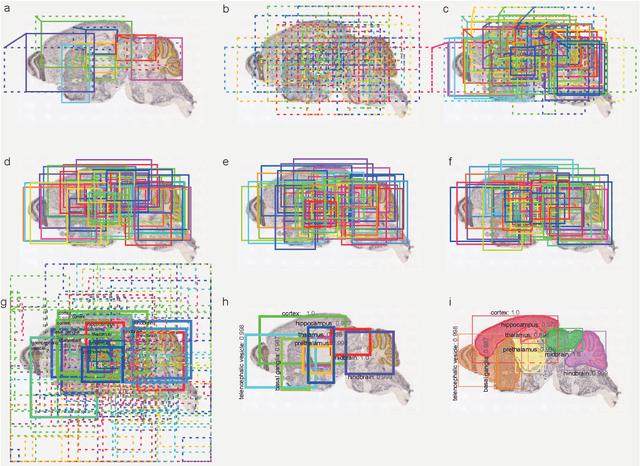
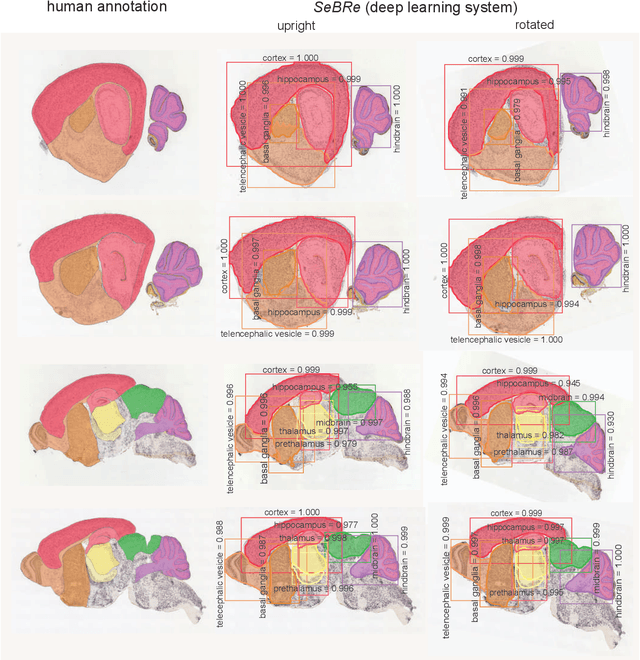
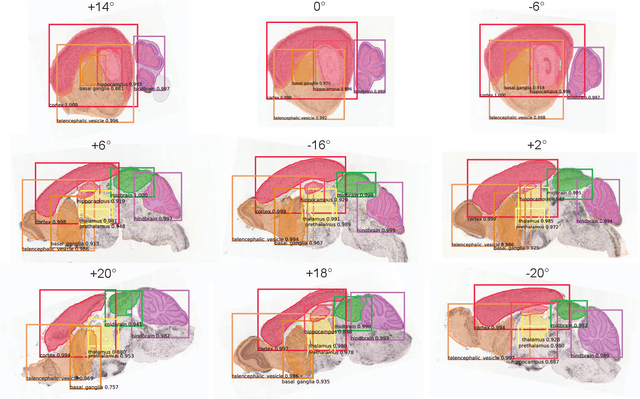
Abstract:To uncover the organizational principles governing the human brain, neuroscientists are in need of developing high-throughput methods that can explore the structure and function of distinct brain regions using animal models. The first step towards this goal is to accurately register the regions of interest in a mouse brain, against a standard reference atlas, with minimum human supervision. The second step is to scale this approach to different animal ages, so as to also allow insights into normal and pathological brain development and aging. We introduce here a fully automated convolutional neural network-based method (SeBRe) for registration through Segmenting Brain Regions of interest in mice at different ages. We demonstrate the validity of our method on different mouse brain post-natal (P) developmental time points, across a range of neuronal markers. Our method outperforms the existing brain registration methods, and provides the minimum mean squared error (MSE) score on a mouse brain dataset. We propose that our deep learning-based registration method can (i) accelerate brain-wide exploration of region-specific changes in brain development and (ii) replace the existing complex brain registration methodology, by simply segmenting brain regions of interest for high-throughput brain-wide analysis.
Exploring Brain-wide Development of Inhibition through Deep Learning
Jul 09, 2018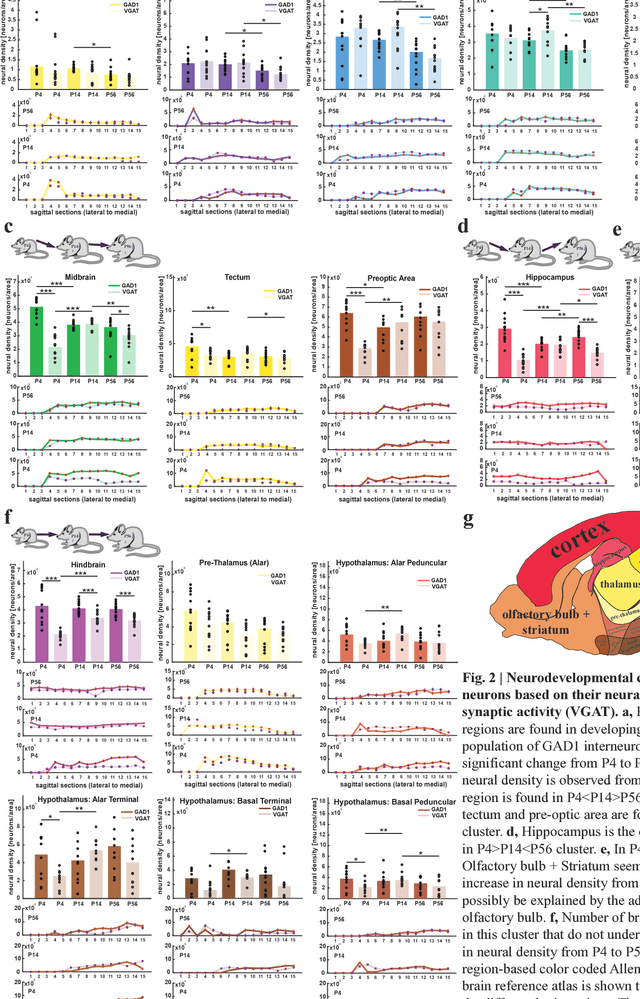
Abstract:We introduce here a fully automated convolutional neural network-based method for brain image processing to Detect Neurons in different brain Regions during Development (DeNeRD). Our method takes a developing mouse brain as input and i) registers the brain sections against a developing mouse reference atlas, ii) detects various types of neurons, and iii) quantifies the neural density in many unique brain regions at different postnatal (P) time points. Our method is invariant to the shape, size and expression of neurons and by using DeNeRD, we compare the brain-wide neural density of all GABAergic neurons in developing brains of ages P4, P14 and P56. We discover and report 6 different clusters of regions in the mouse brain in which GABAergic neurons develop in a differential manner from early age (P4) to adulthood (P56). These clusters reveal key steps of GABAergic cell development that seem to track with the functional development of diverse brain regions as the mouse transitions from a passive receiver of sensory information (<P14) to an active seeker (>P14).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge