Ningsheng Xu
MedQ-Bench: Evaluating and Exploring Medical Image Quality Assessment Abilities in MLLMs
Oct 02, 2025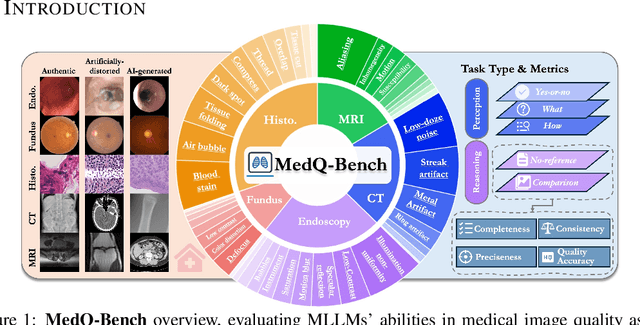
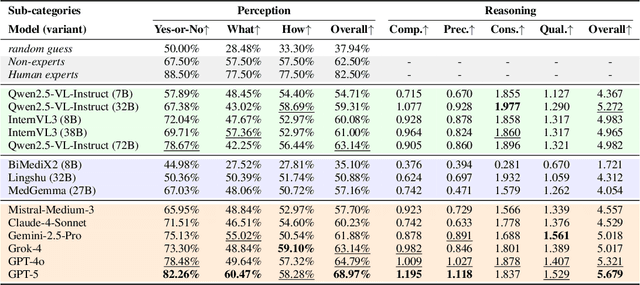
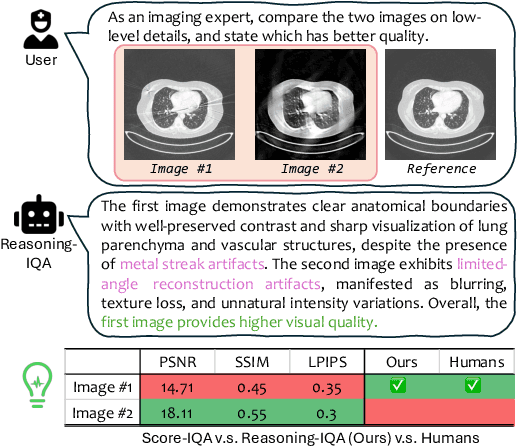
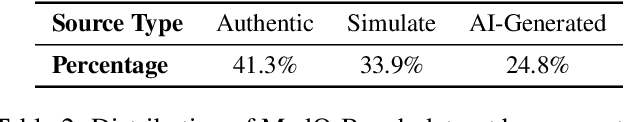
Abstract:Medical Image Quality Assessment (IQA) serves as the first-mile safety gate for clinical AI, yet existing approaches remain constrained by scalar, score-based metrics and fail to reflect the descriptive, human-like reasoning process central to expert evaluation. To address this gap, we introduce MedQ-Bench, a comprehensive benchmark that establishes a perception-reasoning paradigm for language-based evaluation of medical image quality with Multi-modal Large Language Models (MLLMs). MedQ-Bench defines two complementary tasks: (1) MedQ-Perception, which probes low-level perceptual capability via human-curated questions on fundamental visual attributes; and (2) MedQ-Reasoning, encompassing both no-reference and comparison reasoning tasks, aligning model evaluation with human-like reasoning on image quality. The benchmark spans five imaging modalities and over forty quality attributes, totaling 2,600 perceptual queries and 708 reasoning assessments, covering diverse image sources including authentic clinical acquisitions, images with simulated degradations via physics-based reconstructions, and AI-generated images. To evaluate reasoning ability, we propose a multi-dimensional judging protocol that assesses model outputs along four complementary axes. We further conduct rigorous human-AI alignment validation by comparing LLM-based judgement with radiologists. Our evaluation of 14 state-of-the-art MLLMs demonstrates that models exhibit preliminary but unstable perceptual and reasoning skills, with insufficient accuracy for reliable clinical use. These findings highlight the need for targeted optimization of MLLMs in medical IQA. We hope that MedQ-Bench will catalyze further exploration and unlock the untapped potential of MLLMs for medical image quality evaluation.
Sub-diffraction terahertz backpropagation compressive imaging
May 05, 2025Abstract:Terahertz single-pixel imaging (TSPI) has garnered significant attention due to its simplicity and cost-effectiveness. However, the relatively long wavelength of THz waves limits sub-diffraction-scale imaging resolution. Although TSPI technique can achieve sub-wavelength resolution, it requires harsh experimental conditions and time-consuming processes. Here, we propose a sub-diffraction THz backpropagation compressive imaging technique. We illuminate the object with monochromatic continuous-wave THz radiation. The transmitted THz wave is modulated by prearranged patterns generated on the back surface of a 500-{\mu}m-thick silicon wafer, realized through photoexcited carriers using a 532-nm laser. The modulated THz wave is then recorded by a single-element detector. An untrained neural network is employed to iteratively reconstruct the object image with an ultralow compression ratio of 1.5625% under a physical model constraint, thus reducing the long sampling times. To further suppress the diffraction-field effects, embedded with the angular spectrum propagation (ASP) theory to model the diffraction of THz waves during propagation, the network retrieves near-field information from the object, enabling sub-diffraction imaging with a spatial resolution of ~{\lambda}0/7 ({\lambda}0 = 833.3 {\mu}m at 0.36 THz) and eliminating the need for ultrathin photomodulators. This approach provides an efficient solution for advancing THz microscopic imaging and addressing other inverse imaging challenges.
Trustworthy Contrast-enhanced Brain MRI Synthesis
Jul 10, 2024



Abstract:Contrast-enhanced brain MRI (CE-MRI) is a valuable diagnostic technique but may pose health risks and incur high costs. To create safer alternatives, multi-modality medical image translation aims to synthesize CE-MRI images from other available modalities. Although existing methods can generate promising predictions, they still face two challenges, i.e., exhibiting over-confidence and lacking interpretability on predictions. To address the above challenges, this paper introduces TrustI2I, a novel trustworthy method that reformulates multi-to-one medical image translation problem as a multimodal regression problem, aiming to build an uncertainty-aware and reliable system. Specifically, our method leverages deep evidential regression to estimate prediction uncertainties and employs an explicit intermediate and late fusion strategy based on the Mixture of Normal Inverse Gamma (MoNIG) distribution, enhancing both synthesis quality and interpretability. Additionally, we incorporate uncertainty calibration to improve the reliability of uncertainty. Validation on the BraTS2018 dataset demonstrates that our approach surpasses current methods, producing higher-quality images with rational uncertainty estimation.
Digital Twin Brain: a simulation and assimilation platform for whole human brain
Aug 02, 2023Abstract:In this work, we present a computing platform named digital twin brain (DTB) that can simulate spiking neuronal networks of the whole human brain scale and more importantly, a personalized biological brain structure. In comparison to most brain simulations with a homogeneous global structure, we highlight that the sparseness, couplingness and heterogeneity in the sMRI, DTI and PET data of the brain has an essential impact on the efficiency of brain simulation, which is proved from the scaling experiments that the DTB of human brain simulation is communication-intensive and memory-access intensive computing systems rather than computation-intensive. We utilize a number of optimization techniques to balance and integrate the computation loads and communication traffics from the heterogeneous biological structure to the general GPU-based HPC and achieve leading simulation performance for the whole human brain-scaled spiking neuronal networks. On the other hand, the biological structure, equipped with a mesoscopic data assimilation, enables the DTB to investigate brain cognitive function by a reverse-engineering method, which is demonstrated by a digital experiment of visual evaluation on the DTB. Furthermore, we believe that the developing DTB will be a promising powerful platform for a large of research orients including brain-inspiredintelligence, rain disease medicine and brain-machine interface.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge