Runqi Meng
OWT: A Foundational Organ-Wise Tokenization Framework for Medical Imaging
May 08, 2025Abstract:Recent advances in representation learning often rely on holistic, black-box embeddings that entangle multiple semantic components, limiting interpretability and generalization. These issues are especially critical in medical imaging. To address these limitations, we propose an Organ-Wise Tokenization (OWT) framework with a Token Group-based Reconstruction (TGR) training paradigm. Unlike conventional approaches that produce holistic features, OWT explicitly disentangles an image into separable token groups, each corresponding to a distinct organ or semantic entity. Our design ensures each token group encapsulates organ-specific information, boosting interpretability, generalization, and efficiency while allowing fine-grained control in downstream tasks. Experiments on CT and MRI datasets demonstrate the effectiveness of OWT in not only achieving strong image reconstruction and segmentation performance, but also enabling novel semantic-level generation and retrieval applications that are out of reach for standard holistic embedding methods. These findings underscore the potential of OWT as a foundational framework for semantically disentangled representation learning, offering broad scalability and applicability to real-world medical imaging scenarios and beyond.
MAST-Pro: Dynamic Mixture-of-Experts for Adaptive Segmentation of Pan-Tumors with Knowledge-Driven Prompts
Mar 18, 2025Abstract:Accurate tumor segmentation is crucial for cancer diagnosis and treatment. While foundation models have advanced general-purpose segmentation, existing methods still struggle with: (1) limited incorporation of medical priors, (2) imbalance between generic and tumor-specific features, and (3) high computational costs for clinical adaptation. To address these challenges, we propose MAST-Pro (Mixture-of-experts for Adaptive Segmentation of pan-Tumors with knowledge-driven Prompts), a novel framework that integrates dynamic Mixture-of-Experts (D-MoE) and knowledge-driven prompts for pan-tumor segmentation. Specifically, text and anatomical prompts provide domain-specific priors, guiding tumor representation learning, while D-MoE dynamically selects experts to balance generic and tumor-specific feature learning, improving segmentation accuracy across diverse tumor types. To enhance efficiency, we employ Parameter-Efficient Fine-Tuning (PEFT), optimizing MAST-Pro with significantly reduced computational overhead. Experiments on multi-anatomical tumor datasets demonstrate that MAST-Pro outperforms state-of-the-art approaches, achieving up to a 5.20% improvement in average DSC while reducing trainable parameters by 91.04%, without compromising accuracy.
Developing a PET/CT Foundation Model for Cross-Modal Anatomical and Functional Imaging
Mar 04, 2025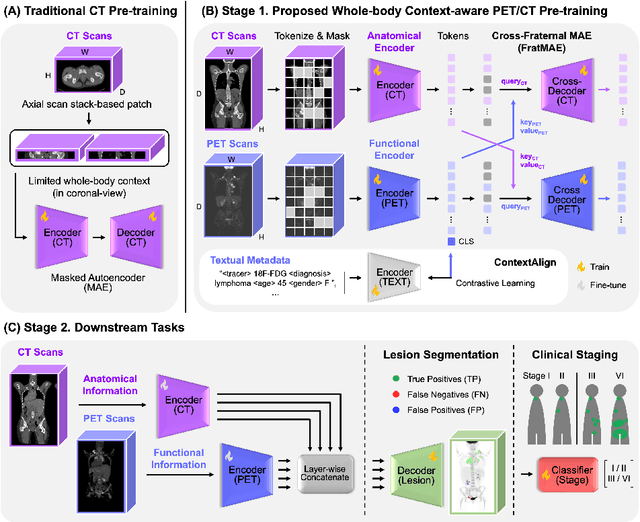
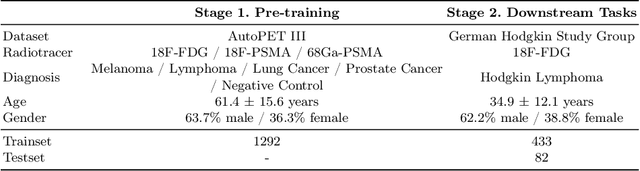
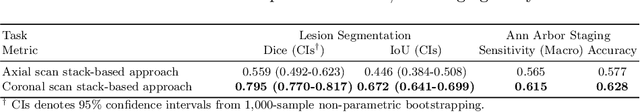
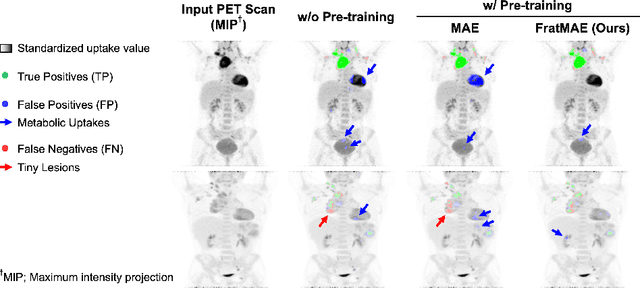
Abstract:In oncology, Positron Emission Tomography-Computed Tomography (PET/CT) is widely used in cancer diagnosis, staging, and treatment monitoring, as it combines anatomical details from CT with functional metabolic activity and molecular marker expression information from PET. However, existing artificial intelligence-driven PET/CT analyses rely predominantly on task-specific models trained from scratch or on limited datasets, limiting their generalizability and robustness. To address this, we propose a foundation model approach specifically designed for multimodal PET/CT imaging. We introduce the Cross-Fraternal Twin Masked Autoencoder (FratMAE), a novel framework that effectively integrates whole-body anatomical and functional or molecular information. FratMAE employs separate Vision Transformer (ViT) encoders for PET and CT scans, along with cross-attention decoders that enable synergistic interactions between modalities during masked autoencoder training. Additionally, it incorporates textual metadata to enhance PET representation learning. By pre-training on PET/CT datasets, FratMAE captures intricate cross-modal relationships and global uptake patterns, achieving superior performance on downstream tasks and demonstrating its potential as a generalizable foundation model.
Knowledge-Guided Prompt Learning for Lifespan Brain MR Image Segmentation
Jul 31, 2024



Abstract:Automatic and accurate segmentation of brain MR images throughout the human lifespan into tissue and structure is crucial for understanding brain development and diagnosing diseases. However, challenges arise from the intricate variations in brain appearance due to rapid early brain development, aging, and disorders, compounded by the limited availability of manually-labeled datasets. In response, we present a two-step segmentation framework employing Knowledge-Guided Prompt Learning (KGPL) for brain MRI. Specifically, we first pre-train segmentation models on large-scale datasets with sub-optimal labels, followed by the incorporation of knowledge-driven embeddings learned from image-text alignment into the models. The introduction of knowledge-wise prompts captures semantic relationships between anatomical variability and biological processes, enabling models to learn structural feature embeddings across diverse age groups. Experimental findings demonstrate the superiority and robustness of our proposed method, particularly noticeable when employing Swin UNETR as the backbone. Our approach achieves average DSC values of 95.17% and 94.19% for brain tissue and structure segmentation, respectively. Our code is available at https://github.com/TL9792/KGPL.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge